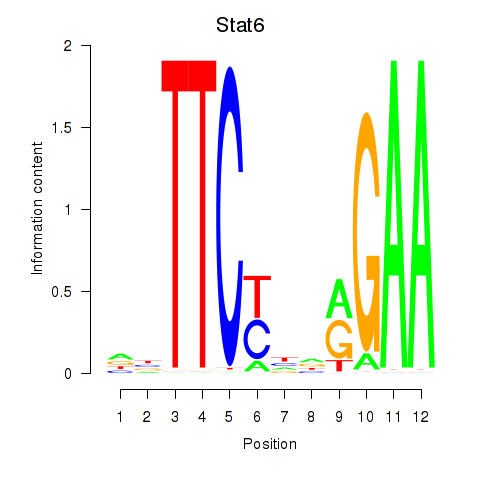
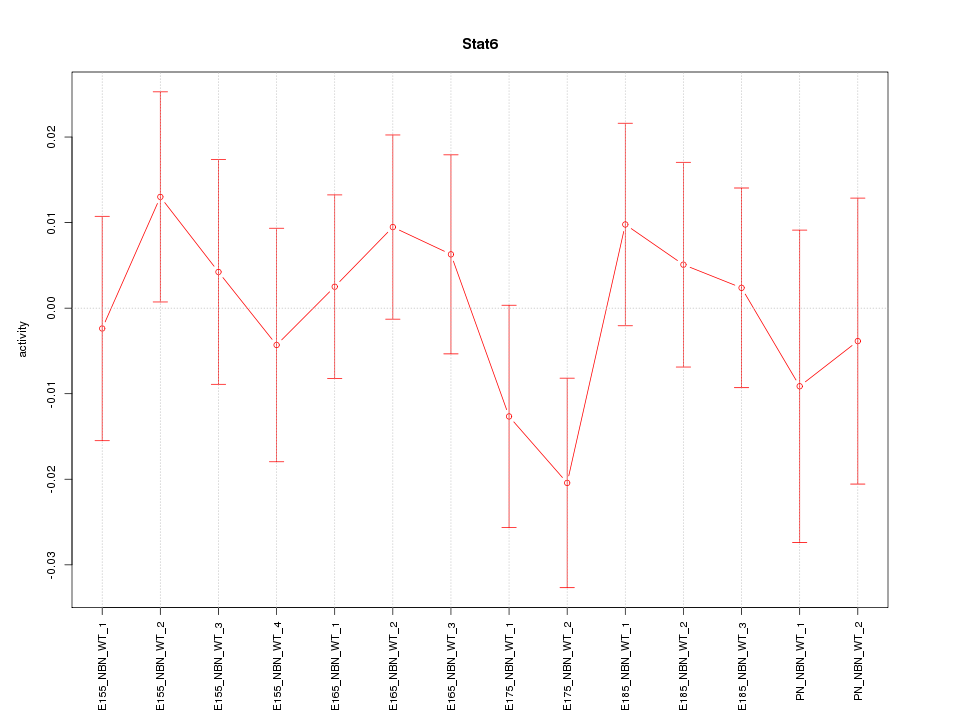
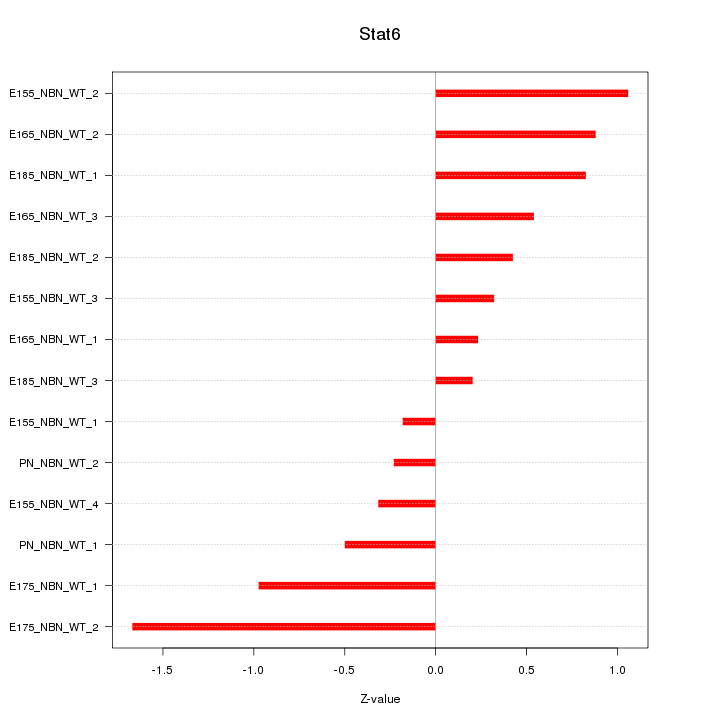
| 0.2 |
0.6 |
GO:0086017 |
renin secretion into blood stream(GO:0002001) cell communication by chemical coupling(GO:0010643) Purkinje myocyte action potential(GO:0086017) regulation of renin secretion into blood stream(GO:1900133) |
| 0.2 |
0.6 |
GO:0070315 |
G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 0.2 |
0.5 |
GO:0070428 |
granuloma formation(GO:0002432) negative regulation of toll-like receptor 3 signaling pathway(GO:0034140) toll-like receptor 5 signaling pathway(GO:0034146) regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) |
| 0.2 |
0.7 |
GO:0051708 |
intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) cytosol to ER transport(GO:0046967) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.2 |
0.5 |
GO:1902524 |
negative regulation of interferon-alpha production(GO:0032687) interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) negative regulation of interferon-beta biosynthetic process(GO:0045358) positive regulation of protein K48-linked ubiquitination(GO:1902524) |
| 0.1 |
0.3 |
GO:0045659 |
regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.1 |
0.3 |
GO:1900740 |
regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.1 |
0.8 |
GO:0030388 |
fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.1 |
0.2 |
GO:0006106 |
fumarate metabolic process(GO:0006106) |
| 0.1 |
0.3 |
GO:0070966 |
nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.1 |
0.5 |
GO:0035726 |
common myeloid progenitor cell proliferation(GO:0035726) |
| 0.1 |
0.2 |
GO:2000383 |
regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.1 |
0.2 |
GO:0070682 |
proteasome regulatory particle assembly(GO:0070682) |
| 0.1 |
0.4 |
GO:1900747 |
negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 |
0.3 |
GO:0016081 |
synaptic vesicle docking(GO:0016081) |
| 0.0 |
0.5 |
GO:0048280 |
vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 |
0.2 |
GO:0036151 |
phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.0 |
0.2 |
GO:0070236 |
negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 |
1.0 |
GO:0095500 |
acetylcholine receptor signaling pathway(GO:0095500) postsynaptic signal transduction(GO:0098926) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.0 |
0.3 |
GO:0000290 |
deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 |
0.1 |
GO:0060423 |
limb joint morphogenesis(GO:0036022) embryonic skeletal limb joint morphogenesis(GO:0036023) negative regulation of mitotic cell cycle, embryonic(GO:0045976) foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) positive regulation of epithelial cell proliferation involved in prostate gland development(GO:0060769) canonical Wnt signaling pathway involved in positive regulation of cardiac outflow tract cell proliferation(GO:0061324) regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901963) regulation of histone demethylase activity (H3-K4 specific)(GO:1904173) |
| 0.0 |
0.5 |
GO:0031507 |
heterochromatin assembly(GO:0031507) |
| 0.0 |
0.4 |
GO:0031274 |
positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 |
0.2 |
GO:0009235 |
cobalamin metabolic process(GO:0009235) |
| 0.0 |
0.5 |
GO:0043011 |
myeloid dendritic cell differentiation(GO:0043011) |
| 0.0 |
0.1 |
GO:0042420 |
dopamine catabolic process(GO:0042420) |
| 0.0 |
0.3 |
GO:0098903 |
regulation of membrane repolarization during action potential(GO:0098903) |
| 0.0 |
0.1 |
GO:0006526 |
arginine biosynthetic process(GO:0006526) |
| 0.0 |
0.1 |
GO:0001803 |
antibody-dependent cellular cytotoxicity(GO:0001788) type III hypersensitivity(GO:0001802) regulation of type III hypersensitivity(GO:0001803) positive regulation of type III hypersensitivity(GO:0001805) |
| 0.0 |
0.1 |
GO:0046684 |
response to pyrethroid(GO:0046684) |
| 0.0 |
0.3 |
GO:0006857 |
oligopeptide transport(GO:0006857) |
| 0.0 |
0.2 |
GO:0006561 |
proline biosynthetic process(GO:0006561) |
| 0.0 |
0.2 |
GO:0090435 |
protein localization to nuclear envelope(GO:0090435) |
| 0.0 |
0.4 |
GO:0000027 |
ribosomal large subunit assembly(GO:0000027) |
| 0.0 |
0.4 |
GO:0018345 |
protein palmitoylation(GO:0018345) |
| 0.0 |
0.3 |
GO:0042036 |
negative regulation of cytokine biosynthetic process(GO:0042036) |
| 0.0 |
0.0 |
GO:2001245 |
regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.0 |
0.1 |
GO:0007379 |
segment specification(GO:0007379) |
| 0.0 |
0.1 |
GO:0003062 |
regulation of heart rate by chemical signal(GO:0003062) |
| 0.0 |
0.0 |
GO:0052428 |
modulation of molecular function in other organism(GO:0044359) negative regulation of molecular function in other organism(GO:0044362) negative regulation of molecular function in other organism involved in symbiotic interaction(GO:0052204) modulation of molecular function in other organism involved in symbiotic interaction(GO:0052205) negative regulation by host of symbiont molecular function(GO:0052405) modification by host of symbiont molecular function(GO:0052428) |
| 0.0 |
0.8 |
GO:0015807 |
L-amino acid transport(GO:0015807) |
| 0.0 |
0.1 |
GO:0006384 |
transcription initiation from RNA polymerase III promoter(GO:0006384) |