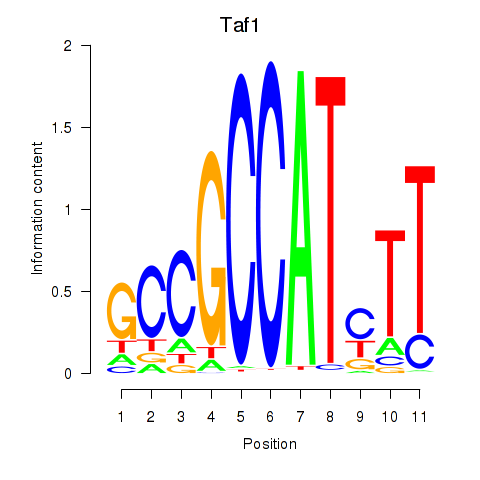
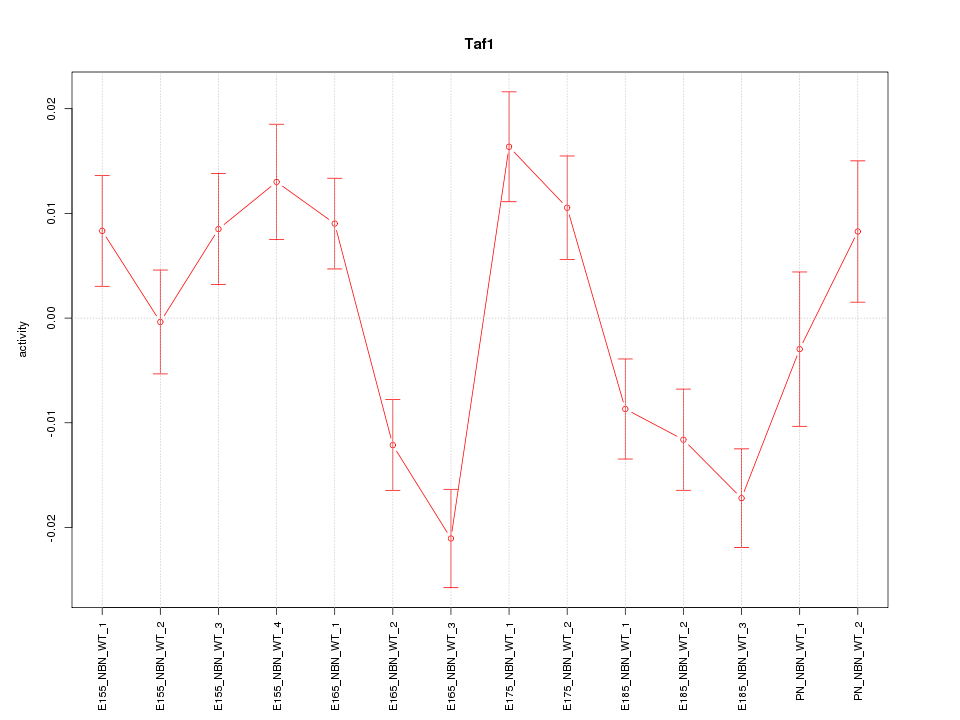
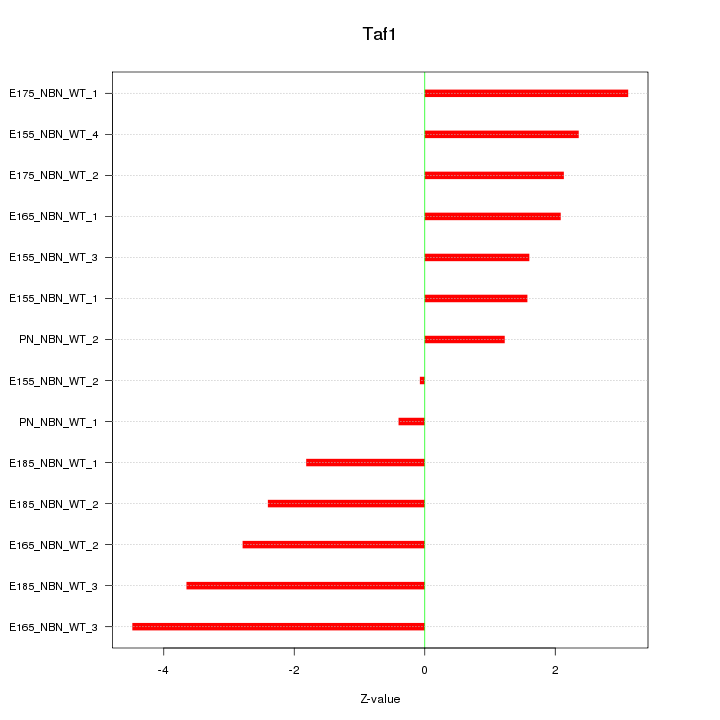
| 1.7 |
6.8 |
GO:0035617 |
stress granule disassembly(GO:0035617) |
| 1.2 |
3.5 |
GO:1902528 |
regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.8 |
3.0 |
GO:2000211 |
regulation of glutamate metabolic process(GO:2000211) |
| 0.8 |
3.8 |
GO:0090234 |
regulation of kinetochore assembly(GO:0090234) |
| 0.7 |
2.9 |
GO:0097393 |
post-embryonic appendage morphogenesis(GO:0035120) post-embryonic limb morphogenesis(GO:0035127) post-embryonic forelimb morphogenesis(GO:0035128) telomeric repeat-containing RNA transcription(GO:0097393) telomeric repeat-containing RNA transcription from RNA pol II promoter(GO:0097394) regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901580) negative regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901581) positive regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901582) |
| 0.7 |
2.8 |
GO:0072362 |
regulation of glycolytic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072362) |
| 0.7 |
2.0 |
GO:0035621 |
ER to Golgi ceramide transport(GO:0035621) ceramide transport(GO:0035627) |
| 0.6 |
0.6 |
GO:0034183 |
negative regulation of maintenance of sister chromatid cohesion(GO:0034092) negative regulation of maintenance of mitotic sister chromatid cohesion(GO:0034183) maintenance of mitotic sister chromatid cohesion, telomeric(GO:0099403) mitotic sister chromatid cohesion, telomeric(GO:0099404) regulation of maintenance of mitotic sister chromatid cohesion, telomeric(GO:1904907) negative regulation of maintenance of mitotic sister chromatid cohesion, telomeric(GO:1904908) |
| 0.6 |
1.7 |
GO:1990481 |
snoRNA guided rRNA pseudouridine synthesis(GO:0000454) snRNA pseudouridine synthesis(GO:0031120) mRNA pseudouridine synthesis(GO:1990481) |
| 0.6 |
2.8 |
GO:0030242 |
pexophagy(GO:0030242) |
| 0.6 |
2.8 |
GO:0061086 |
negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.5 |
1.6 |
GO:1902037 |
negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 0.5 |
2.1 |
GO:0071596 |
ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.5 |
2.1 |
GO:2001032 |
regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.5 |
1.5 |
GO:1901254 |
positive regulation of intracellular transport of viral material(GO:1901254) |
| 0.5 |
2.3 |
GO:1900095 |
regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.5 |
2.3 |
GO:0010032 |
meiotic chromosome condensation(GO:0010032) |
| 0.4 |
1.3 |
GO:0001827 |
inner cell mass cell fate commitment(GO:0001827) |
| 0.4 |
1.8 |
GO:0019042 |
viral latency(GO:0019042) |
| 0.4 |
4.3 |
GO:1903608 |
protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.4 |
2.1 |
GO:0034421 |
post-translational protein acetylation(GO:0034421) |
| 0.4 |
1.7 |
GO:0097494 |
regulation of vesicle size(GO:0097494) |
| 0.4 |
2.5 |
GO:0021773 |
striatal medium spiny neuron differentiation(GO:0021773) |
| 0.4 |
2.4 |
GO:0030643 |
cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.4 |
1.2 |
GO:1900275 |
negative regulation of phospholipase C activity(GO:1900275) regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900736) |
| 0.4 |
1.1 |
GO:0019858 |
cytosine metabolic process(GO:0019858) |
| 0.4 |
1.1 |
GO:0032240 |
RNA import into nucleus(GO:0006404) mRNA export from nucleus in response to heat stress(GO:0031990) negative regulation of nucleobase-containing compound transport(GO:0032240) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.4 |
1.1 |
GO:0045204 |
MAPK export from nucleus(GO:0045204) |
| 0.4 |
2.1 |
GO:0097368 |
establishment of Sertoli cell barrier(GO:0097368) |
| 0.3 |
1.7 |
GO:1901843 |
positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.3 |
1.0 |
GO:0032915 |
positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 0.3 |
2.6 |
GO:0071557 |
histone H3-K27 demethylation(GO:0071557) |
| 0.3 |
1.2 |
GO:0021698 |
cerebellar cortex structural organization(GO:0021698) |
| 0.3 |
0.9 |
GO:0060821 |
inactivation of X chromosome by DNA methylation(GO:0060821) |
| 0.3 |
1.7 |
GO:0051660 |
cortical microtubule organization(GO:0043622) establishment of centrosome localization(GO:0051660) |
| 0.3 |
0.8 |
GO:0034184 |
positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.3 |
1.7 |
GO:1900246 |
positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.3 |
2.3 |
GO:0018344 |
protein geranylgeranylation(GO:0018344) |
| 0.3 |
1.9 |
GO:0016584 |
nucleosome positioning(GO:0016584) |
| 0.3 |
0.8 |
GO:0042275 |
error-free postreplication DNA repair(GO:0042275) |
| 0.3 |
0.8 |
GO:2000686 |
regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.3 |
0.8 |
GO:1904017 |
response to Thyroglobulin triiodothyronine(GO:1904016) cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.3 |
2.3 |
GO:0030854 |
positive regulation of granulocyte differentiation(GO:0030854) |
| 0.2 |
2.5 |
GO:2000051 |
negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.2 |
0.7 |
GO:0019074 |
viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.2 |
0.7 |
GO:0019085 |
early viral transcription(GO:0019085) |
| 0.2 |
0.7 |
GO:0070358 |
actin polymerization-dependent cell motility(GO:0070358) |
| 0.2 |
0.9 |
GO:0034454 |
microtubule anchoring at centrosome(GO:0034454) |
| 0.2 |
2.9 |
GO:0060009 |
Sertoli cell development(GO:0060009) |
| 0.2 |
1.3 |
GO:0021631 |
optic nerve morphogenesis(GO:0021631) |
| 0.2 |
0.6 |
GO:0031590 |
wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.2 |
0.6 |
GO:0031627 |
telomeric loop formation(GO:0031627) |
| 0.2 |
1.0 |
GO:0002036 |
regulation of L-glutamate transport(GO:0002036) |
| 0.2 |
0.6 |
GO:0032916 |
positive regulation of transforming growth factor beta3 production(GO:0032916) |
| 0.2 |
1.4 |
GO:0035507 |
regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.2 |
0.6 |
GO:0043323 |
positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.2 |
1.6 |
GO:0010793 |
regulation of mRNA export from nucleus(GO:0010793) regulation of ribonucleoprotein complex localization(GO:2000197) |
| 0.2 |
0.8 |
GO:0070318 |
positive regulation of G0 to G1 transition(GO:0070318) |
| 0.2 |
0.9 |
GO:0001778 |
plasma membrane repair(GO:0001778) |
| 0.2 |
0.9 |
GO:0006369 |
termination of RNA polymerase II transcription(GO:0006369) |
| 0.2 |
0.2 |
GO:0000375 |
RNA splicing, via transesterification reactions(GO:0000375) |
| 0.2 |
0.6 |
GO:0009405 |
pathogenesis(GO:0009405) |
| 0.2 |
1.7 |
GO:0033148 |
positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.2 |
0.7 |
GO:0097709 |
connective tissue replacement involved in inflammatory response wound healing(GO:0002248) connective tissue replacement(GO:0097709) |
| 0.2 |
0.6 |
GO:1901420 |
pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) cellular response to testosterone stimulus(GO:0071394) negative regulation of response to alcohol(GO:1901420) positive regulation of macrophage apoptotic process(GO:2000111) |
| 0.2 |
0.6 |
GO:0061535 |
glutamate secretion, neurotransmission(GO:0061535) |
| 0.2 |
0.5 |
GO:0007228 |
positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.2 |
0.5 |
GO:0002669 |
positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) protein K29-linked ubiquitination(GO:0035519) |
| 0.2 |
0.9 |
GO:0060050 |
positive regulation of protein glycosylation(GO:0060050) |
| 0.2 |
0.8 |
GO:1903361 |
protein localization to basolateral plasma membrane(GO:1903361) |
| 0.2 |
0.7 |
GO:0035521 |
monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.2 |
1.7 |
GO:0032876 |
negative regulation of DNA endoreduplication(GO:0032876) |
| 0.2 |
0.7 |
GO:1902626 |
assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.2 |
0.5 |
GO:0043379 |
memory T cell differentiation(GO:0043379) |
| 0.2 |
0.7 |
GO:0030950 |
establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.2 |
2.4 |
GO:0060736 |
prostate gland growth(GO:0060736) |
| 0.2 |
1.3 |
GO:0031936 |
negative regulation of chromatin silencing(GO:0031936) |
| 0.2 |
0.8 |
GO:0060024 |
rhythmic synaptic transmission(GO:0060024) |
| 0.2 |
1.1 |
GO:0017196 |
N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.2 |
0.6 |
GO:0035616 |
histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.2 |
0.8 |
GO:2000301 |
negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.2 |
0.5 |
GO:0006435 |
threonyl-tRNA aminoacylation(GO:0006435) |
| 0.1 |
0.4 |
GO:0070103 |
interleukin-4-mediated signaling pathway(GO:0035771) tyrosine phosphorylation of Stat6 protein(GO:0042505) regulation of tyrosine phosphorylation of Stat6 protein(GO:0042525) regulation of interleukin-6-mediated signaling pathway(GO:0070103) negative regulation of interleukin-6-mediated signaling pathway(GO:0070104) |
| 0.1 |
0.7 |
GO:2000623 |
regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.1 |
0.7 |
GO:1990928 |
response to amino acid starvation(GO:1990928) |
| 0.1 |
0.7 |
GO:0046601 |
positive regulation of centriole replication(GO:0046601) |
| 0.1 |
1.7 |
GO:0031268 |
pseudopodium organization(GO:0031268) |
| 0.1 |
2.0 |
GO:0001731 |
formation of translation preinitiation complex(GO:0001731) |
| 0.1 |
1.0 |
GO:0006620 |
posttranslational protein targeting to membrane(GO:0006620) |
| 0.1 |
0.1 |
GO:0045876 |
positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.1 |
0.4 |
GO:0097500 |
receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.1 |
1.1 |
GO:0090169 |
regulation of spindle assembly(GO:0090169) regulation of mitotic spindle assembly(GO:1901673) |
| 0.1 |
0.7 |
GO:0070966 |
nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.1 |
2.5 |
GO:0045653 |
negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.1 |
1.5 |
GO:0045974 |
miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.1 |
1.2 |
GO:0046339 |
diacylglycerol metabolic process(GO:0046339) |
| 0.1 |
0.9 |
GO:0071699 |
olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.1 |
1.0 |
GO:0080182 |
histone H3-K4 trimethylation(GO:0080182) |
| 0.1 |
0.4 |
GO:0042998 |
positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 0.1 |
1.4 |
GO:0010650 |
positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.1 |
0.4 |
GO:0018076 |
N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.1 |
0.4 |
GO:0070934 |
CRD-mediated mRNA stabilization(GO:0070934) |
| 0.1 |
1.0 |
GO:0034773 |
histone H4-K20 trimethylation(GO:0034773) |
| 0.1 |
1.0 |
GO:0035871 |
protein K11-linked deubiquitination(GO:0035871) |
| 0.1 |
0.6 |
GO:0060158 |
phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.1 |
0.7 |
GO:0032380 |
regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.1 |
0.4 |
GO:0021691 |
cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.1 |
0.9 |
GO:0007076 |
mitotic chromosome condensation(GO:0007076) |
| 0.1 |
0.7 |
GO:0002710 |
negative regulation of T cell mediated immunity(GO:0002710) |
| 0.1 |
0.7 |
GO:0048254 |
snoRNA localization(GO:0048254) |
| 0.1 |
0.4 |
GO:2000574 |
regulation of microtubule motor activity(GO:2000574) |
| 0.1 |
0.5 |
GO:0010993 |
regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 0.1 |
0.7 |
GO:0045218 |
zonula adherens maintenance(GO:0045218) |
| 0.1 |
0.5 |
GO:2001274 |
negative regulation of glucose import in response to insulin stimulus(GO:2001274) |
| 0.1 |
0.3 |
GO:0035526 |
retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.1 |
0.9 |
GO:0000395 |
mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 |
2.0 |
GO:0031442 |
positive regulation of mRNA 3'-end processing(GO:0031442) |
| 0.1 |
2.4 |
GO:0051457 |
maintenance of protein location in nucleus(GO:0051457) |
| 0.1 |
1.8 |
GO:0015693 |
magnesium ion transport(GO:0015693) |
| 0.1 |
2.7 |
GO:0045722 |
positive regulation of gluconeogenesis(GO:0045722) |
| 0.1 |
0.7 |
GO:0031848 |
protection from non-homologous end joining at telomere(GO:0031848) |
| 0.1 |
0.3 |
GO:0097278 |
virion attachment to host cell(GO:0019062) adhesion of symbiont to host cell(GO:0044650) complement-dependent cytotoxicity(GO:0097278) |
| 0.1 |
0.3 |
GO:0019482 |
beta-alanine metabolic process(GO:0019482) |
| 0.1 |
0.6 |
GO:0090245 |
axis elongation involved in somitogenesis(GO:0090245) |
| 0.1 |
0.7 |
GO:0060046 |
regulation of acrosome reaction(GO:0060046) |
| 0.1 |
0.7 |
GO:0015074 |
DNA integration(GO:0015074) |
| 0.1 |
0.4 |
GO:0070425 |
negative regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070425) negative regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070433) |
| 0.1 |
0.2 |
GO:0070213 |
protein auto-ADP-ribosylation(GO:0070213) |
| 0.1 |
0.1 |
GO:0034382 |
chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) |
| 0.1 |
1.0 |
GO:0036371 |
protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.1 |
1.2 |
GO:2000381 |
negative regulation of mesoderm development(GO:2000381) |
| 0.1 |
1.3 |
GO:0043508 |
negative regulation of JUN kinase activity(GO:0043508) |
| 0.1 |
0.5 |
GO:0071267 |
amino acid salvage(GO:0043102) L-methionine biosynthetic process(GO:0071265) L-methionine salvage(GO:0071267) |
| 0.1 |
0.5 |
GO:0071894 |
histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.1 |
0.7 |
GO:2000601 |
positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 |
0.3 |
GO:0030862 |
regulation of polarized epithelial cell differentiation(GO:0030860) positive regulation of polarized epithelial cell differentiation(GO:0030862) |
| 0.1 |
0.4 |
GO:2001045 |
closure of optic fissure(GO:0061386) negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 |
1.0 |
GO:0042118 |
endothelial cell activation(GO:0042118) |
| 0.1 |
1.4 |
GO:1904424 |
regulation of GTP binding(GO:1904424) |
| 0.1 |
0.6 |
GO:1902410 |
mitotic cytokinetic process(GO:1902410) |
| 0.1 |
0.8 |
GO:2000270 |
negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.1 |
1.1 |
GO:0000059 |
protein import into nucleus, docking(GO:0000059) |
| 0.1 |
0.3 |
GO:1902915 |
negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.1 |
0.3 |
GO:2000616 |
negative regulation of histone H3-K9 acetylation(GO:2000616) |
| 0.1 |
0.4 |
GO:0090370 |
negative regulation of cholesterol efflux(GO:0090370) |
| 0.1 |
0.3 |
GO:0070537 |
histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.1 |
1.1 |
GO:0036120 |
cellular response to platelet-derived growth factor stimulus(GO:0036120) |
| 0.1 |
1.4 |
GO:0045579 |
positive regulation of B cell differentiation(GO:0045579) |
| 0.1 |
2.5 |
GO:0007099 |
centriole replication(GO:0007099) |
| 0.1 |
0.3 |
GO:0006421 |
asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.1 |
0.5 |
GO:0006379 |
mRNA cleavage(GO:0006379) |
| 0.1 |
0.4 |
GO:0048007 |
antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.1 |
1.2 |
GO:0034501 |
protein localization to kinetochore(GO:0034501) |
| 0.1 |
0.7 |
GO:0055089 |
fatty acid homeostasis(GO:0055089) |
| 0.1 |
1.2 |
GO:0034453 |
microtubule anchoring(GO:0034453) |
| 0.1 |
0.3 |
GO:1903265 |
positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.1 |
0.3 |
GO:0010890 |
positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.1 |
0.3 |
GO:0043987 |
histone H3-S10 phosphorylation(GO:0043987) histone H3-S28 phosphorylation(GO:0043988) |
| 0.1 |
0.2 |
GO:0035622 |
intrahepatic bile duct development(GO:0035622) |
| 0.1 |
0.3 |
GO:0006546 |
glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.1 |
0.7 |
GO:1902065 |
response to L-glutamate(GO:1902065) |
| 0.1 |
0.5 |
GO:1902866 |
regulation of retina development in camera-type eye(GO:1902866) |
| 0.1 |
2.2 |
GO:0000413 |
protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 |
0.3 |
GO:0070829 |
heterochromatin maintenance(GO:0070829) |
| 0.1 |
0.9 |
GO:0002098 |
tRNA wobble uridine modification(GO:0002098) |
| 0.1 |
0.2 |
GO:0070973 |
protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 |
0.6 |
GO:0006398 |
mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.1 |
0.2 |
GO:0045617 |
negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.1 |
0.5 |
GO:0051461 |
regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 0.1 |
0.7 |
GO:0060363 |
cranial suture morphogenesis(GO:0060363) |
| 0.1 |
0.2 |
GO:1903998 |
regulation of eating behavior(GO:1903998) |
| 0.1 |
0.9 |
GO:0070914 |
UV-damage excision repair(GO:0070914) |
| 0.1 |
0.1 |
GO:0008626 |
granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.1 |
0.5 |
GO:0035459 |
cargo loading into vesicle(GO:0035459) |
| 0.1 |
1.4 |
GO:0006415 |
translational termination(GO:0006415) |
| 0.1 |
1.5 |
GO:1903861 |
regulation of dendrite extension(GO:1903859) positive regulation of dendrite extension(GO:1903861) |
| 0.1 |
0.4 |
GO:0035405 |
histone-threonine phosphorylation(GO:0035405) |
| 0.1 |
0.9 |
GO:0060746 |
parental behavior(GO:0060746) |
| 0.1 |
0.5 |
GO:0006621 |
protein retention in ER lumen(GO:0006621) |
| 0.1 |
0.2 |
GO:0016598 |
protein arginylation(GO:0016598) |
| 0.1 |
1.5 |
GO:0044030 |
regulation of DNA methylation(GO:0044030) |
| 0.1 |
0.4 |
GO:0006344 |
optic cup morphogenesis involved in camera-type eye development(GO:0002072) maintenance of chromatin silencing(GO:0006344) |
| 0.1 |
0.4 |
GO:0036515 |
serotonergic neuron axon guidance(GO:0036515) |
| 0.1 |
0.3 |
GO:0006543 |
glutamine catabolic process(GO:0006543) |
| 0.1 |
1.9 |
GO:0018345 |
protein palmitoylation(GO:0018345) |
| 0.1 |
2.2 |
GO:0031648 |
protein destabilization(GO:0031648) |
| 0.1 |
0.4 |
GO:0070072 |
vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.1 |
0.1 |
GO:1903378 |
positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.1 |
0.7 |
GO:0031468 |
nuclear envelope reassembly(GO:0031468) |
| 0.1 |
0.3 |
GO:0071105 |
response to interleukin-11(GO:0071105) osteoclast fusion(GO:0072675) |
| 0.1 |
0.4 |
GO:0051534 |
negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.1 |
0.6 |
GO:0000712 |
resolution of meiotic recombination intermediates(GO:0000712) |
| 0.1 |
0.1 |
GO:0060489 |
establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) dichotomous subdivision of terminal units involved in lung branching(GO:0060448) orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.1 |
0.6 |
GO:0071763 |
nuclear membrane organization(GO:0071763) |
| 0.1 |
0.3 |
GO:0051189 |
Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.1 |
0.1 |
GO:0031086 |
nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.1 |
0.2 |
GO:0048211 |
Golgi vesicle docking(GO:0048211) |
| 0.1 |
0.6 |
GO:0055090 |
acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.1 |
1.1 |
GO:0099517 |
anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.1 |
1.1 |
GO:0007020 |
microtubule nucleation(GO:0007020) |
| 0.1 |
0.6 |
GO:2001256 |
regulation of store-operated calcium entry(GO:2001256) |
| 0.1 |
1.5 |
GO:0008053 |
mitochondrial fusion(GO:0008053) |
| 0.1 |
0.4 |
GO:0051127 |
positive regulation of actin nucleation(GO:0051127) |
| 0.1 |
0.6 |
GO:0001672 |
regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.1 |
0.3 |
GO:0031293 |
membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.1 |
1.0 |
GO:0008090 |
retrograde axonal transport(GO:0008090) |
| 0.1 |
0.2 |
GO:0060729 |
preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) intestinal epithelial structure maintenance(GO:0060729) |
| 0.1 |
0.9 |
GO:0001522 |
pseudouridine synthesis(GO:0001522) |
| 0.1 |
1.8 |
GO:0006376 |
mRNA splice site selection(GO:0006376) |
| 0.1 |
0.6 |
GO:0016926 |
protein desumoylation(GO:0016926) |
| 0.1 |
0.6 |
GO:0021942 |
radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.1 |
0.5 |
GO:0006013 |
mannose metabolic process(GO:0006013) |
| 0.1 |
0.2 |
GO:0046836 |
glycolipid transport(GO:0046836) |
| 0.1 |
0.2 |
GO:0032790 |
ribosome disassembly(GO:0032790) |
| 0.1 |
0.5 |
GO:0031937 |
positive regulation of chromatin silencing(GO:0031937) |
| 0.1 |
0.2 |
GO:0009446 |
putrescine biosynthetic process(GO:0009446) |
| 0.1 |
0.6 |
GO:0007175 |
negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 |
0.2 |
GO:0048388 |
endosomal lumen acidification(GO:0048388) synaptic vesicle lumen acidification(GO:0097401) |
| 0.1 |
0.5 |
GO:2000780 |
negative regulation of double-strand break repair(GO:2000780) |
| 0.1 |
0.4 |
GO:0043619 |
regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.1 |
0.3 |
GO:0048069 |
eye pigmentation(GO:0048069) |
| 0.1 |
0.2 |
GO:0044154 |
histone H3-K14 acetylation(GO:0044154) |
| 0.1 |
0.6 |
GO:0002176 |
male germ cell proliferation(GO:0002176) |
| 0.1 |
0.3 |
GO:0060017 |
parathyroid gland development(GO:0060017) |
| 0.1 |
2.2 |
GO:0017145 |
stem cell division(GO:0017145) |
| 0.1 |
0.3 |
GO:1900042 |
positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.0 |
0.1 |
GO:0018171 |
peptidyl-cysteine oxidation(GO:0018171) |
| 0.0 |
0.1 |
GO:0006436 |
tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.0 |
0.8 |
GO:0048520 |
positive regulation of behavior(GO:0048520) |
| 0.0 |
1.7 |
GO:0006904 |
vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 |
0.5 |
GO:0032926 |
negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 |
0.1 |
GO:0006578 |
amino-acid betaine biosynthetic process(GO:0006578) |
| 0.0 |
0.5 |
GO:0007250 |
activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 |
0.3 |
GO:0048549 |
positive regulation of pinocytosis(GO:0048549) |
| 0.0 |
0.3 |
GO:0060674 |
placenta blood vessel development(GO:0060674) |
| 0.0 |
0.7 |
GO:0010640 |
regulation of platelet-derived growth factor receptor signaling pathway(GO:0010640) |
| 0.0 |
0.6 |
GO:0016973 |
poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 |
1.7 |
GO:0007032 |
endosome organization(GO:0007032) |
| 0.0 |
1.7 |
GO:0048009 |
insulin-like growth factor receptor signaling pathway(GO:0048009) |
| 0.0 |
0.2 |
GO:0043461 |
proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.0 |
0.5 |
GO:0015677 |
copper ion import(GO:0015677) |
| 0.0 |
0.1 |
GO:0030210 |
heparin biosynthetic process(GO:0030210) |
| 0.0 |
0.1 |
GO:0008594 |
photoreceptor cell morphogenesis(GO:0008594) |
| 0.0 |
0.9 |
GO:0001574 |
ganglioside biosynthetic process(GO:0001574) |
| 0.0 |
0.4 |
GO:0060766 |
negative regulation of androgen receptor signaling pathway(GO:0060766) |
| 0.0 |
0.7 |
GO:0071108 |
protein K48-linked deubiquitination(GO:0071108) |
| 0.0 |
0.3 |
GO:0031053 |
primary miRNA processing(GO:0031053) |
| 0.0 |
0.5 |
GO:0031340 |
positive regulation of vesicle fusion(GO:0031340) |
| 0.0 |
0.5 |
GO:0099500 |
synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.0 |
0.4 |
GO:0071549 |
response to dexamethasone(GO:0071548) cellular response to dexamethasone stimulus(GO:0071549) |
| 0.0 |
0.2 |
GO:0010826 |
negative regulation of centrosome duplication(GO:0010826) negative regulation of centrosome cycle(GO:0046606) |
| 0.0 |
0.2 |
GO:0071955 |
recycling endosome to Golgi transport(GO:0071955) |
| 0.0 |
0.5 |
GO:0016558 |
protein import into peroxisome matrix(GO:0016558) |
| 0.0 |
0.2 |
GO:0000022 |
mitotic spindle elongation(GO:0000022) spindle midzone assembly(GO:0051255) mitotic spindle midzone assembly(GO:0051256) |
| 0.0 |
0.3 |
GO:0090073 |
negative regulation of telomere maintenance via telomerase(GO:0032211) positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 |
0.5 |
GO:0006471 |
protein ADP-ribosylation(GO:0006471) |
| 0.0 |
0.0 |
GO:0006287 |
base-excision repair, gap-filling(GO:0006287) |
| 0.0 |
0.2 |
GO:1902035 |
positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.0 |
0.4 |
GO:0007064 |
mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 |
0.3 |
GO:0042754 |
negative regulation of circadian rhythm(GO:0042754) |
| 0.0 |
0.4 |
GO:2000114 |
regulation of establishment of cell polarity(GO:2000114) |
| 0.0 |
0.5 |
GO:0060416 |
response to growth hormone(GO:0060416) |
| 0.0 |
0.3 |
GO:0006817 |
phosphate ion transport(GO:0006817) |
| 0.0 |
0.2 |
GO:0060689 |
cell differentiation involved in salivary gland development(GO:0060689) |
| 0.0 |
0.7 |
GO:0006359 |
regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 |
0.6 |
GO:0043984 |
histone H4-K16 acetylation(GO:0043984) |
| 0.0 |
1.1 |
GO:0016601 |
Rac protein signal transduction(GO:0016601) |
| 0.0 |
1.5 |
GO:0008045 |
motor neuron axon guidance(GO:0008045) |
| 0.0 |
5.4 |
GO:0042787 |
protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.0 |
0.1 |
GO:0009414 |
response to water deprivation(GO:0009414) |
| 0.0 |
0.1 |
GO:0071169 |
establishment of protein localization to chromatin(GO:0071169) |
| 0.0 |
0.6 |
GO:0043968 |
histone H2A acetylation(GO:0043968) |
| 0.0 |
0.2 |
GO:0006297 |
nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.0 |
0.3 |
GO:0045176 |
apical protein localization(GO:0045176) |
| 0.0 |
0.8 |
GO:0010955 |
negative regulation of protein processing(GO:0010955) negative regulation of protein maturation(GO:1903318) |
| 0.0 |
2.8 |
GO:0021954 |
central nervous system neuron development(GO:0021954) |
| 0.0 |
0.3 |
GO:0045671 |
negative regulation of osteoclast differentiation(GO:0045671) |
| 0.0 |
0.3 |
GO:0010800 |
positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 |
0.3 |
GO:0006878 |
cellular copper ion homeostasis(GO:0006878) |
| 0.0 |
0.2 |
GO:1904217 |
regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) regulation of serine C-palmitoyltransferase activity(GO:1904220) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.0 |
0.1 |
GO:0035520 |
monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 |
0.1 |
GO:0090237 |
regulation of arachidonic acid secretion(GO:0090237) positive regulation of arachidonic acid secretion(GO:0090238) |
| 0.0 |
0.1 |
GO:0042373 |
vitamin K metabolic process(GO:0042373) |
| 0.0 |
0.4 |
GO:0034724 |
DNA replication-independent nucleosome assembly(GO:0006336) DNA replication-independent nucleosome organization(GO:0034724) |
| 0.0 |
0.1 |
GO:0071168 |
protein localization to chromatin(GO:0071168) |
| 0.0 |
0.2 |
GO:0060124 |
positive regulation of growth hormone secretion(GO:0060124) |
| 0.0 |
1.8 |
GO:0010923 |
negative regulation of phosphatase activity(GO:0010923) |
| 0.0 |
0.2 |
GO:0046325 |
negative regulation of glucose import(GO:0046325) |
| 0.0 |
0.1 |
GO:0045448 |
regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.0 |
0.2 |
GO:0042985 |
negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.0 |
1.0 |
GO:0031663 |
lipopolysaccharide-mediated signaling pathway(GO:0031663) |
| 0.0 |
0.1 |
GO:0046477 |
glycosylceramide catabolic process(GO:0046477) |
| 0.0 |
0.9 |
GO:0006879 |
cellular iron ion homeostasis(GO:0006879) |
| 0.0 |
0.6 |
GO:0048268 |
clathrin coat assembly(GO:0048268) |
| 0.0 |
0.2 |
GO:0030007 |
cellular potassium ion homeostasis(GO:0030007) |
| 0.0 |
0.9 |
GO:0036075 |
endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 |
0.2 |
GO:0070989 |
oxidative demethylation(GO:0070989) |
| 0.0 |
0.1 |
GO:2000973 |
regulation of pro-B cell differentiation(GO:2000973) |
| 0.0 |
0.3 |
GO:0051292 |
nuclear pore complex assembly(GO:0051292) |
| 0.0 |
0.6 |
GO:0035909 |
aorta morphogenesis(GO:0035909) |
| 0.0 |
1.3 |
GO:0002224 |
toll-like receptor signaling pathway(GO:0002224) |
| 0.0 |
0.2 |
GO:0048681 |
negative regulation of axon regeneration(GO:0048681) |
| 0.0 |
0.2 |
GO:0042487 |
regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.0 |
0.1 |
GO:0006867 |
asparagine transport(GO:0006867) glutamine transport(GO:0006868) |
| 0.0 |
0.2 |
GO:0003356 |
regulation of cilium movement(GO:0003352) regulation of cilium beat frequency(GO:0003356) |
| 0.0 |
0.3 |
GO:1990403 |
embryonic brain development(GO:1990403) |
| 0.0 |
0.6 |
GO:0003170 |
heart valve development(GO:0003170) |
| 0.0 |
0.3 |
GO:0060693 |
regulation of branching involved in salivary gland morphogenesis(GO:0060693) |
| 0.0 |
0.3 |
GO:2000637 |
positive regulation of gene silencing by miRNA(GO:2000637) |
| 0.0 |
0.1 |
GO:0018206 |
peptidyl-methionine modification(GO:0018206) |
| 0.0 |
0.9 |
GO:0060711 |
labyrinthine layer development(GO:0060711) |
| 0.0 |
0.1 |
GO:0033147 |
negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 |
0.1 |
GO:0048102 |
autophagic cell death(GO:0048102) |
| 0.0 |
0.4 |
GO:0046827 |
positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 |
0.3 |
GO:0071578 |
zinc II ion transmembrane import(GO:0071578) |
| 0.0 |
0.6 |
GO:0090102 |
cochlea development(GO:0090102) |
| 0.0 |
0.9 |
GO:0007492 |
endoderm development(GO:0007492) |
| 0.0 |
0.2 |
GO:0033631 |
cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.0 |
0.1 |
GO:0051315 |
attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.0 |
0.8 |
GO:0043268 |
positive regulation of potassium ion transport(GO:0043268) |
| 0.0 |
0.1 |
GO:0072520 |
seminiferous tubule development(GO:0072520) |
| 0.0 |
0.1 |
GO:0048496 |
maintenance of organ identity(GO:0048496) |
| 0.0 |
0.1 |
GO:0048199 |
vesicle targeting, to, from or within Golgi(GO:0048199) |
| 0.0 |
0.2 |
GO:0006390 |
transcription from mitochondrial promoter(GO:0006390) |
| 0.0 |
0.2 |
GO:0010457 |
centriole-centriole cohesion(GO:0010457) |
| 0.0 |
0.3 |
GO:0006388 |
tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 |
0.2 |
GO:0000083 |
regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 |
0.1 |
GO:0045721 |
negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 |
0.1 |
GO:0000188 |
inactivation of MAPK activity(GO:0000188) |
| 0.0 |
0.7 |
GO:0019228 |
neuronal action potential(GO:0019228) |
| 0.0 |
0.2 |
GO:0000479 |
endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000479) |
| 0.0 |
0.2 |
GO:0043278 |
response to isoquinoline alkaloid(GO:0014072) response to morphine(GO:0043278) |
| 0.0 |
0.1 |
GO:0051152 |
positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 |
0.6 |
GO:0006378 |
mRNA polyadenylation(GO:0006378) |
| 0.0 |
0.1 |
GO:0046084 |
adenine metabolic process(GO:0046083) adenine biosynthetic process(GO:0046084) |
| 0.0 |
0.2 |
GO:0045647 |
negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 |
0.1 |
GO:0036506 |
maintenance of unfolded protein(GO:0036506) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.0 |
0.1 |
GO:0006020 |
inositol metabolic process(GO:0006020) |
| 0.0 |
0.1 |
GO:1901838 |
positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.0 |
0.2 |
GO:0080009 |
mRNA methylation(GO:0080009) |
| 0.0 |
0.3 |
GO:0006884 |
cell volume homeostasis(GO:0006884) |
| 0.0 |
0.4 |
GO:0045494 |
photoreceptor cell maintenance(GO:0045494) |
| 0.0 |
0.3 |
GO:1902043 |
positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.0 |
0.2 |
GO:0048671 |
negative regulation of collateral sprouting(GO:0048671) |
| 0.0 |
0.4 |
GO:0043392 |
negative regulation of DNA binding(GO:0043392) |
| 0.0 |
0.1 |
GO:0008088 |
axo-dendritic transport(GO:0008088) |
| 0.0 |
0.4 |
GO:0045197 |
establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.0 |
0.3 |
GO:0007205 |
protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 |
0.1 |
GO:0019254 |
carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 |
1.5 |
GO:0002244 |
hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.0 |
0.1 |
GO:0008343 |
adult feeding behavior(GO:0008343) |
| 0.0 |
0.0 |
GO:0042264 |
peptidyl-aspartic acid modification(GO:0018197) peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 |
0.1 |
GO:0018065 |
protein-cofactor linkage(GO:0018065) |
| 0.0 |
0.3 |
GO:0032008 |
positive regulation of TOR signaling(GO:0032008) |
| 0.0 |
0.0 |
GO:0052428 |
modulation of molecular function in other organism(GO:0044359) negative regulation of molecular function in other organism(GO:0044362) negative regulation of molecular function in other organism involved in symbiotic interaction(GO:0052204) modulation of molecular function in other organism involved in symbiotic interaction(GO:0052205) negative regulation by host of symbiont molecular function(GO:0052405) modification by host of symbiont molecular function(GO:0052428) |
| 0.0 |
0.4 |
GO:0007520 |
myoblast fusion(GO:0007520) |
| 0.0 |
0.0 |
GO:0097499 |
protein localization to nonmotile primary cilium(GO:0097499) |
| 0.0 |
0.2 |
GO:0006654 |
phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.0 |
0.5 |
GO:0035418 |
protein localization to synapse(GO:0035418) |
| 0.0 |
0.1 |
GO:0014051 |
gamma-aminobutyric acid secretion(GO:0014051) |
| 0.0 |
0.1 |
GO:1901409 |
regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 |
0.1 |
GO:0030836 |
positive regulation of actin filament depolymerization(GO:0030836) |
| 0.0 |
0.2 |
GO:0070534 |
protein K63-linked ubiquitination(GO:0070534) |
| 0.0 |
0.1 |
GO:0048318 |
axial mesoderm development(GO:0048318) |
| 0.0 |
0.2 |
GO:0008340 |
determination of adult lifespan(GO:0008340) |
| 0.0 |
0.0 |
GO:0015882 |
L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.0 |
0.3 |
GO:0033539 |
fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 |
0.1 |
GO:0035493 |
SNARE complex assembly(GO:0035493) |
| 0.0 |
0.1 |
GO:0007031 |
peroxisome organization(GO:0007031) |
| 0.0 |
0.3 |
GO:0046579 |
positive regulation of Ras protein signal transduction(GO:0046579) |
| 0.0 |
0.4 |
GO:0045668 |
negative regulation of osteoblast differentiation(GO:0045668) |
| 0.0 |
0.0 |
GO:1902512 |
positive regulation of apoptotic DNA fragmentation(GO:1902512) positive regulation of DNA catabolic process(GO:1903626) |
| 0.0 |
0.1 |
GO:0031584 |
activation of phospholipase D activity(GO:0031584) |
| 0.0 |
0.1 |
GO:1902775 |
mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 |
0.5 |
GO:0000380 |
alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.0 |
0.1 |
GO:0090141 |
positive regulation of mitochondrial fission(GO:0090141) |
| 0.0 |
0.5 |
GO:0007631 |
feeding behavior(GO:0007631) |
| 0.0 |
0.0 |
GO:0046831 |
regulation of RNA export from nucleus(GO:0046831) |
| 0.0 |
0.4 |
GO:0006936 |
muscle contraction(GO:0006936) |