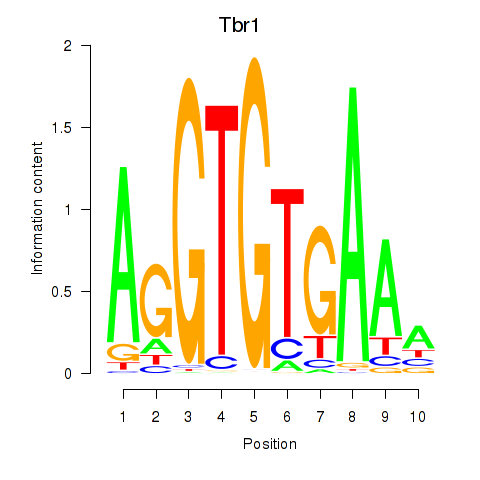
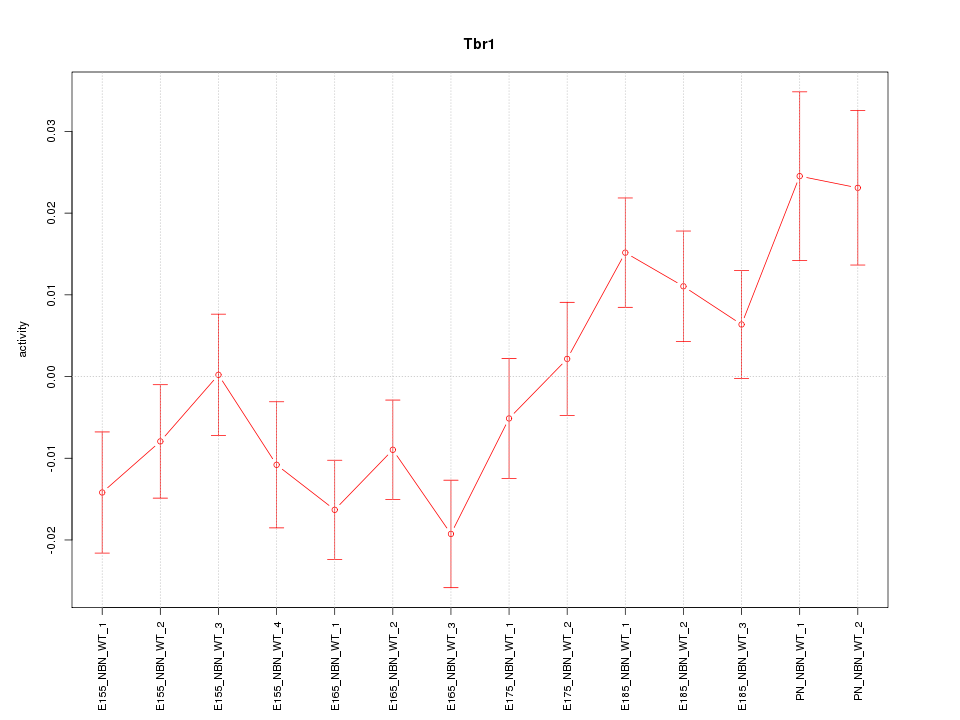
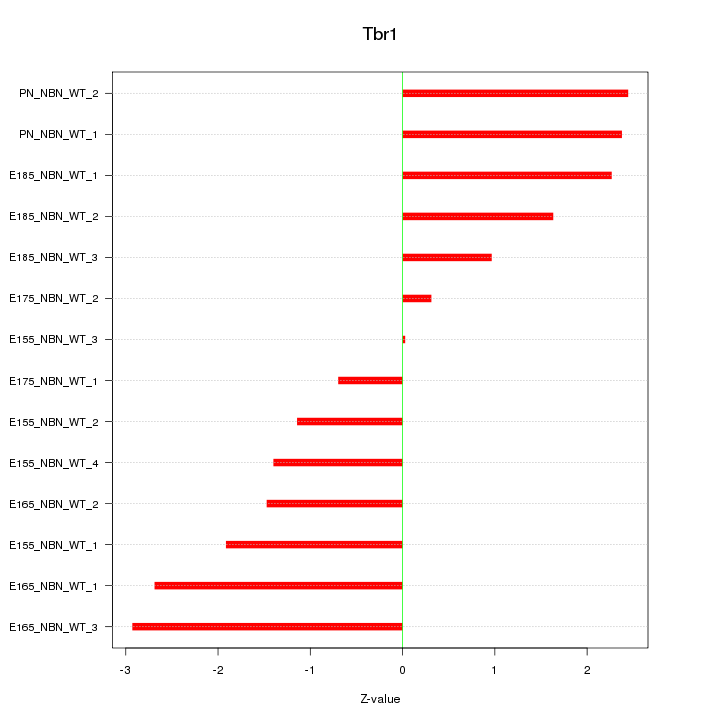
| 4.4 |
13.1 |
GO:0021893 |
cerebral cortex GABAergic interneuron fate commitment(GO:0021893) |
| 1.5 |
4.4 |
GO:0061537 |
glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 1.4 |
4.2 |
GO:0006116 |
NADH oxidation(GO:0006116) |
| 1.1 |
3.4 |
GO:2001139 |
negative regulation of postsynaptic membrane organization(GO:1901627) negative regulation of dendritic spine maintenance(GO:1902951) negative regulation of phospholipid efflux(GO:1902999) regulation of lipid transport across blood brain barrier(GO:1903000) negative regulation of lipid transport across blood brain barrier(GO:1903001) positive regulation of lipid transport across blood brain barrier(GO:1903002) negative regulation of phospholipid transport(GO:2001139) |
| 1.1 |
3.2 |
GO:2000562 |
negative regulation of interferon-gamma secretion(GO:1902714) negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 1.1 |
3.2 |
GO:0036233 |
glycine import(GO:0036233) |
| 1.0 |
5.2 |
GO:0048245 |
eosinophil chemotaxis(GO:0048245) |
| 1.0 |
3.1 |
GO:0001803 |
antibody-dependent cellular cytotoxicity(GO:0001788) type III hypersensitivity(GO:0001802) regulation of type III hypersensitivity(GO:0001803) positive regulation of type III hypersensitivity(GO:0001805) |
| 0.9 |
3.8 |
GO:0035934 |
corticosterone secretion(GO:0035934) |
| 0.9 |
2.7 |
GO:0048818 |
positive regulation of hair follicle maturation(GO:0048818) positive regulation of catagen(GO:0051795) regulation of epithelial to mesenchymal transition involved in endocardial cushion formation(GO:1905005) |
| 0.9 |
2.7 |
GO:0019343 |
cysteine biosynthetic process via cystathionine(GO:0019343) cysteine biosynthetic process(GO:0019344) |
| 0.8 |
4.2 |
GO:2001204 |
regulation of osteoclast development(GO:2001204) |
| 0.8 |
0.8 |
GO:1900673 |
olefin metabolic process(GO:1900673) |
| 0.8 |
2.4 |
GO:0030421 |
defecation(GO:0030421) |
| 0.7 |
3.7 |
GO:0030091 |
protein repair(GO:0030091) |
| 0.7 |
2.1 |
GO:0003245 |
cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.7 |
2.1 |
GO:2000983 |
regulation of ATP citrate synthase activity(GO:2000983) negative regulation of ATP citrate synthase activity(GO:2000984) |
| 0.7 |
3.3 |
GO:0002457 |
T cell antigen processing and presentation(GO:0002457) |
| 0.6 |
1.9 |
GO:1902358 |
sulfate transmembrane transport(GO:1902358) |
| 0.6 |
1.8 |
GO:0003278 |
apoptotic process involved in heart morphogenesis(GO:0003278) |
| 0.6 |
2.2 |
GO:0032610 |
interleukin-1 alpha production(GO:0032610) |
| 0.5 |
1.1 |
GO:1905065 |
positive regulation of vascular smooth muscle cell differentiation(GO:1905065) |
| 0.5 |
2.1 |
GO:2000048 |
negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.5 |
1.4 |
GO:0060672 |
epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.5 |
1.4 |
GO:0050859 |
negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.4 |
1.3 |
GO:1901628 |
positive regulation of postsynaptic membrane organization(GO:1901628) |
| 0.4 |
1.7 |
GO:2001032 |
regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.4 |
1.1 |
GO:0014738 |
regulation of muscle hyperplasia(GO:0014738) muscle hyperplasia(GO:0014900) |
| 0.4 |
1.5 |
GO:0051725 |
protein de-ADP-ribosylation(GO:0051725) |
| 0.4 |
1.8 |
GO:2000504 |
positive regulation of blood vessel remodeling(GO:2000504) |
| 0.4 |
1.1 |
GO:1903984 |
positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.4 |
1.4 |
GO:0001907 |
killing by symbiont of host cells(GO:0001907) induction of programmed cell death(GO:0012502) disruption by symbiont of host cell(GO:0044004) positive regulation of apoptotic process in other organism(GO:0044533) positive regulation by symbiont of host programmed cell death(GO:0052042) positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052330) positive regulation by organism of apoptotic process in other organism involved in symbiotic interaction(GO:0052501) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.4 |
0.7 |
GO:0045054 |
constitutive secretory pathway(GO:0045054) |
| 0.4 |
1.1 |
GO:0002430 |
complement receptor mediated signaling pathway(GO:0002430) |
| 0.3 |
1.4 |
GO:0061010 |
gall bladder development(GO:0061010) |
| 0.3 |
2.1 |
GO:0034316 |
negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.3 |
1.0 |
GO:2000501 |
natural killer cell chemotaxis(GO:0035747) regulation of natural killer cell chemotaxis(GO:2000501) |
| 0.3 |
5.9 |
GO:0060134 |
prepulse inhibition(GO:0060134) |
| 0.3 |
1.6 |
GO:1901679 |
nucleotide transmembrane transport(GO:1901679) |
| 0.3 |
1.0 |
GO:0045014 |
detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) detection of glucose(GO:0051594) |
| 0.3 |
2.5 |
GO:0009181 |
purine nucleoside diphosphate catabolic process(GO:0009137) purine ribonucleoside diphosphate catabolic process(GO:0009181) |
| 0.3 |
0.9 |
GO:0035621 |
ER to Golgi ceramide transport(GO:0035621) ceramide transport(GO:0035627) |
| 0.3 |
4.4 |
GO:0010447 |
response to acidic pH(GO:0010447) |
| 0.3 |
1.2 |
GO:0006933 |
negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.3 |
0.9 |
GO:0090526 |
response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.3 |
0.9 |
GO:2000271 |
positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.3 |
2.3 |
GO:0015670 |
carbon dioxide transport(GO:0015670) |
| 0.3 |
0.8 |
GO:1903232 |
melanosome assembly(GO:1903232) |
| 0.3 |
1.1 |
GO:0048633 |
positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.3 |
0.8 |
GO:1990481 |
snoRNA guided rRNA pseudouridine synthesis(GO:0000454) snRNA pseudouridine synthesis(GO:0031120) mRNA pseudouridine synthesis(GO:1990481) |
| 0.3 |
1.1 |
GO:0032788 |
saturated monocarboxylic acid metabolic process(GO:0032788) unsaturated monocarboxylic acid metabolic process(GO:0032789) |
| 0.3 |
0.8 |
GO:0035633 |
maintenance of blood-brain barrier(GO:0035633) |
| 0.3 |
1.8 |
GO:0090336 |
positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.3 |
3.3 |
GO:0043252 |
sodium-independent organic anion transport(GO:0043252) |
| 0.3 |
0.5 |
GO:0040031 |
snRNA modification(GO:0040031) |
| 0.3 |
1.5 |
GO:2001185 |
regulation of CD8-positive, alpha-beta T cell activation(GO:2001185) |
| 0.2 |
0.7 |
GO:0002586 |
positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002586) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) |
| 0.2 |
2.2 |
GO:2000194 |
regulation of female gonad development(GO:2000194) |
| 0.2 |
1.0 |
GO:0071726 |
response to diacyl bacterial lipopeptide(GO:0071724) cellular response to diacyl bacterial lipopeptide(GO:0071726) |
| 0.2 |
0.7 |
GO:0007521 |
muscle cell fate determination(GO:0007521) mammary placode formation(GO:0060596) |
| 0.2 |
1.2 |
GO:0051315 |
attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.2 |
0.9 |
GO:0072137 |
condensed mesenchymal cell proliferation(GO:0072137) |
| 0.2 |
0.9 |
GO:0018343 |
protein farnesylation(GO:0018343) negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.2 |
1.1 |
GO:0035262 |
gonad morphogenesis(GO:0035262) |
| 0.2 |
1.2 |
GO:0060648 |
mammary gland bud morphogenesis(GO:0060648) |
| 0.2 |
1.4 |
GO:0035948 |
positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) regulation of cellular ketone metabolic process by positive regulation of transcription from RNA polymerase II promoter(GO:0072366) |
| 0.2 |
1.2 |
GO:0050855 |
regulation of B cell receptor signaling pathway(GO:0050855) positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.2 |
0.8 |
GO:0060161 |
positive regulation of dopamine receptor signaling pathway(GO:0060161) |
| 0.2 |
2.6 |
GO:0045647 |
negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.2 |
0.4 |
GO:0007386 |
compartment pattern specification(GO:0007386) |
| 0.2 |
3.1 |
GO:0006953 |
acute-phase response(GO:0006953) |
| 0.2 |
1.4 |
GO:0006297 |
nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.2 |
2.9 |
GO:0035994 |
response to muscle stretch(GO:0035994) |
| 0.2 |
1.5 |
GO:0048752 |
semicircular canal morphogenesis(GO:0048752) |
| 0.2 |
3.0 |
GO:0042428 |
serotonin metabolic process(GO:0042428) |
| 0.2 |
2.8 |
GO:0021860 |
pyramidal neuron development(GO:0021860) |
| 0.2 |
1.1 |
GO:0045218 |
zonula adherens maintenance(GO:0045218) |
| 0.2 |
0.5 |
GO:0003349 |
epicardium-derived cardiac endothelial cell differentiation(GO:0003349) |
| 0.2 |
0.7 |
GO:0007128 |
meiotic prophase I(GO:0007128) prophase(GO:0051324) meiotic cell cycle phase(GO:0098762) meiosis I cell cycle phase(GO:0098764) |
| 0.2 |
0.7 |
GO:1903215 |
negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.2 |
2.2 |
GO:0010457 |
centriole-centriole cohesion(GO:0010457) |
| 0.2 |
0.8 |
GO:0050747 |
positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.1 |
0.6 |
GO:0071449 |
cellular response to lipid hydroperoxide(GO:0071449) |
| 0.1 |
0.4 |
GO:0009812 |
flavonoid metabolic process(GO:0009812) flavonoid biosynthetic process(GO:0009813) flavonoid glucuronidation(GO:0052696) |
| 0.1 |
0.9 |
GO:0045591 |
positive regulation of regulatory T cell differentiation(GO:0045591) |
| 0.1 |
0.4 |
GO:1902528 |
regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.1 |
1.0 |
GO:0000022 |
mitotic spindle elongation(GO:0000022) mitotic spindle midzone assembly(GO:0051256) |
| 0.1 |
1.0 |
GO:0043951 |
negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.1 |
0.7 |
GO:2000483 |
negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.1 |
1.0 |
GO:0006686 |
sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 |
0.8 |
GO:0010528 |
regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.1 |
6.5 |
GO:0043666 |
regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.1 |
0.8 |
GO:1904715 |
negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 0.1 |
1.1 |
GO:0035735 |
intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.1 |
1.8 |
GO:0043402 |
glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.1 |
0.8 |
GO:0034227 |
tRNA thio-modification(GO:0034227) |
| 0.1 |
0.5 |
GO:0006049 |
UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.1 |
0.4 |
GO:0032474 |
otolith morphogenesis(GO:0032474) |
| 0.1 |
4.0 |
GO:0070306 |
lens fiber cell differentiation(GO:0070306) |
| 0.1 |
0.5 |
GO:0006041 |
glucosamine metabolic process(GO:0006041) |
| 0.1 |
0.5 |
GO:0000707 |
meiotic DNA recombinase assembly(GO:0000707) resolution of recombination intermediates(GO:0071139) |
| 0.1 |
0.4 |
GO:0036015 |
response to interleukin-3(GO:0036015) cellular response to interleukin-3(GO:0036016) |
| 0.1 |
0.9 |
GO:0036506 |
maintenance of unfolded protein(GO:0036506) protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.1 |
0.3 |
GO:0035526 |
retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.1 |
0.7 |
GO:0045605 |
negative regulation of epidermal cell differentiation(GO:0045605) |
| 0.1 |
1.6 |
GO:0002495 |
antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.1 |
0.5 |
GO:0046684 |
response to pyrethroid(GO:0046684) |
| 0.1 |
0.9 |
GO:0044130 |
negative regulation of growth of symbiont in host(GO:0044130) negative regulation of growth of symbiont involved in interaction with host(GO:0044146) |
| 0.1 |
1.2 |
GO:0060538 |
skeletal muscle tissue development(GO:0007519) skeletal muscle organ development(GO:0060538) |
| 0.1 |
0.8 |
GO:0015865 |
purine nucleotide transport(GO:0015865) |
| 0.1 |
0.6 |
GO:0002317 |
plasma cell differentiation(GO:0002317) |
| 0.1 |
1.5 |
GO:0021891 |
olfactory bulb interneuron development(GO:0021891) |
| 0.1 |
0.4 |
GO:0014819 |
regulation of skeletal muscle contraction(GO:0014819) |
| 0.1 |
0.6 |
GO:0045654 |
positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.1 |
0.8 |
GO:0071493 |
cellular response to UV-B(GO:0071493) replicative senescence(GO:0090399) |
| 0.1 |
2.0 |
GO:1903861 |
regulation of dendrite extension(GO:1903859) positive regulation of dendrite extension(GO:1903861) |
| 0.1 |
0.3 |
GO:0044028 |
DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.1 |
0.3 |
GO:0045204 |
MAPK export from nucleus(GO:0045204) |
| 0.1 |
0.3 |
GO:0018094 |
protein polyglycylation(GO:0018094) |
| 0.1 |
2.3 |
GO:0015985 |
energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 |
0.2 |
GO:0046101 |
hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 0.1 |
0.6 |
GO:0032534 |
regulation of microvillus assembly(GO:0032534) |
| 0.1 |
0.4 |
GO:1901837 |
negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.1 |
0.5 |
GO:1903352 |
ornithine transport(GO:0015822) L-ornithine transmembrane transport(GO:1903352) |
| 0.1 |
0.5 |
GO:0006172 |
ADP biosynthetic process(GO:0006172) |
| 0.1 |
0.5 |
GO:0048254 |
snoRNA localization(GO:0048254) |
| 0.1 |
1.4 |
GO:0032757 |
positive regulation of interleukin-8 production(GO:0032757) |
| 0.1 |
0.4 |
GO:0030242 |
pexophagy(GO:0030242) |
| 0.1 |
0.6 |
GO:2000786 |
positive regulation of autophagosome assembly(GO:2000786) |
| 0.1 |
0.5 |
GO:0060923 |
muscle cell fate commitment(GO:0042693) cardiac muscle cell fate commitment(GO:0060923) |
| 0.1 |
0.4 |
GO:0015803 |
branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.1 |
0.3 |
GO:0035521 |
monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.1 |
0.3 |
GO:0006478 |
peptidyl-tyrosine sulfation(GO:0006478) |
| 0.1 |
0.2 |
GO:0031652 |
positive regulation of heat generation(GO:0031652) |
| 0.1 |
0.9 |
GO:0070995 |
NADPH oxidation(GO:0070995) |
| 0.1 |
0.4 |
GO:0061086 |
negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.1 |
0.2 |
GO:0033561 |
regulation of water loss via skin(GO:0033561) establishment of skin barrier(GO:0061436) |
| 0.1 |
0.7 |
GO:0090051 |
negative regulation of cell migration involved in sprouting angiogenesis(GO:0090051) |
| 0.1 |
1.0 |
GO:0072010 |
renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.1 |
0.5 |
GO:0070458 |
detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 0.1 |
0.2 |
GO:2000569 |
positive regulation of interleukin-17 production(GO:0032740) T-helper 2 cell activation(GO:0035712) positive regulation of T-helper 17 type immune response(GO:2000318) regulation of T-helper 2 cell activation(GO:2000569) positive regulation of T-helper 2 cell activation(GO:2000570) |
| 0.1 |
1.5 |
GO:0048791 |
calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 |
0.3 |
GO:2000383 |
regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.1 |
0.8 |
GO:1902459 |
positive regulation of stem cell population maintenance(GO:1902459) |
| 0.1 |
0.9 |
GO:0042219 |
cellular modified amino acid catabolic process(GO:0042219) |
| 0.1 |
0.5 |
GO:0010908 |
regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.1 |
1.7 |
GO:0009409 |
response to cold(GO:0009409) |
| 0.1 |
0.4 |
GO:0015074 |
DNA integration(GO:0015074) |
| 0.1 |
0.4 |
GO:2000781 |
positive regulation of double-strand break repair(GO:2000781) |
| 0.1 |
1.8 |
GO:0018345 |
protein palmitoylation(GO:0018345) |
| 0.1 |
0.9 |
GO:0050832 |
defense response to fungus(GO:0050832) |
| 0.1 |
0.4 |
GO:0009992 |
cellular water homeostasis(GO:0009992) |
| 0.1 |
0.2 |
GO:0072675 |
osteoclast fusion(GO:0072675) |
| 0.1 |
0.2 |
GO:0031038 |
myosin II filament organization(GO:0031038) regulation of myosin II filament organization(GO:0043519) |
| 0.1 |
0.2 |
GO:0070814 |
hydrogen sulfide biosynthetic process(GO:0070814) |
| 0.1 |
0.7 |
GO:2000406 |
positive regulation of T cell migration(GO:2000406) |
| 0.1 |
0.4 |
GO:0014883 |
transition between fast and slow fiber(GO:0014883) |
| 0.1 |
1.4 |
GO:0050873 |
brown fat cell differentiation(GO:0050873) |
| 0.1 |
0.4 |
GO:0006177 |
GMP biosynthetic process(GO:0006177) |
| 0.1 |
2.3 |
GO:0031114 |
negative regulation of microtubule depolymerization(GO:0007026) regulation of microtubule depolymerization(GO:0031114) |
| 0.1 |
0.4 |
GO:0051013 |
microtubule severing(GO:0051013) |
| 0.1 |
0.8 |
GO:0043589 |
skin morphogenesis(GO:0043589) |
| 0.1 |
1.4 |
GO:0071108 |
protein K48-linked deubiquitination(GO:0071108) |
| 0.1 |
0.7 |
GO:0060391 |
positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.1 |
0.2 |
GO:0043504 |
mitochondrial DNA repair(GO:0043504) |
| 0.1 |
0.4 |
GO:0007220 |
Notch receptor processing(GO:0007220) |
| 0.1 |
0.3 |
GO:0006564 |
L-serine biosynthetic process(GO:0006564) |
| 0.1 |
1.6 |
GO:0050853 |
B cell receptor signaling pathway(GO:0050853) |
| 0.1 |
0.4 |
GO:0035562 |
negative regulation of chromatin binding(GO:0035562) |
| 0.1 |
0.1 |
GO:0001951 |
intestinal D-glucose absorption(GO:0001951) terminal web assembly(GO:1902896) |
| 0.1 |
2.4 |
GO:0046676 |
negative regulation of insulin secretion(GO:0046676) |
| 0.1 |
0.9 |
GO:2000096 |
positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.1 |
0.1 |
GO:0010958 |
regulation of amino acid import(GO:0010958) regulation of L-arginine import(GO:0010963) |
| 0.1 |
0.2 |
GO:0034476 |
U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.1 |
0.8 |
GO:0034453 |
microtubule anchoring(GO:0034453) |
| 0.1 |
0.5 |
GO:0019800 |
peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 |
1.0 |
GO:1900153 |
regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.0 |
0.1 |
GO:0097623 |
potassium ion export across plasma membrane(GO:0097623) |
| 0.0 |
2.1 |
GO:0050709 |
negative regulation of protein secretion(GO:0050709) |
| 0.0 |
0.2 |
GO:0015871 |
choline transport(GO:0015871) |
| 0.0 |
1.8 |
GO:0051310 |
metaphase plate congression(GO:0051310) |
| 0.0 |
0.6 |
GO:0045954 |
positive regulation of natural killer cell mediated immunity(GO:0002717) positive regulation of natural killer cell mediated cytotoxicity(GO:0045954) |
| 0.0 |
0.3 |
GO:1903894 |
regulation of IRE1-mediated unfolded protein response(GO:1903894) |
| 0.0 |
0.3 |
GO:0018344 |
protein geranylgeranylation(GO:0018344) |
| 0.0 |
0.6 |
GO:0006120 |
mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 |
1.7 |
GO:0001919 |
regulation of receptor recycling(GO:0001919) |
| 0.0 |
0.2 |
GO:0010359 |
regulation of anion channel activity(GO:0010359) |
| 0.0 |
0.2 |
GO:0060158 |
phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 |
0.4 |
GO:0009072 |
aromatic amino acid family metabolic process(GO:0009072) |
| 0.0 |
0.3 |
GO:0044331 |
cell-cell adhesion mediated by cadherin(GO:0044331) |
| 0.0 |
0.2 |
GO:0010025 |
wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.0 |
0.3 |
GO:0030539 |
male genitalia development(GO:0030539) |
| 0.0 |
0.3 |
GO:0032464 |
positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 |
0.4 |
GO:0007250 |
activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 |
0.5 |
GO:0042573 |
retinoic acid metabolic process(GO:0042573) |
| 0.0 |
0.2 |
GO:0006842 |
tricarboxylic acid transport(GO:0006842) succinate transport(GO:0015744) citrate transport(GO:0015746) |
| 0.0 |
0.1 |
GO:0015882 |
L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.0 |
0.0 |
GO:0070366 |
regulation of hepatocyte differentiation(GO:0070366) |
| 0.0 |
1.6 |
GO:0042771 |
intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 |
0.2 |
GO:0006384 |
transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 |
0.7 |
GO:2000001 |
regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 |
0.8 |
GO:0070232 |
regulation of T cell apoptotic process(GO:0070232) |
| 0.0 |
0.3 |
GO:0048643 |
positive regulation of skeletal muscle tissue development(GO:0048643) |
| 0.0 |
1.0 |
GO:0060441 |
epithelial tube branching involved in lung morphogenesis(GO:0060441) |
| 0.0 |
1.3 |
GO:0000387 |
spliceosomal snRNP assembly(GO:0000387) |
| 0.0 |
0.1 |
GO:0046598 |
positive regulation of viral entry into host cell(GO:0046598) |
| 0.0 |
0.1 |
GO:0006668 |
sphinganine-1-phosphate metabolic process(GO:0006668) |
| 0.0 |
1.5 |
GO:0045744 |
negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.0 |
0.5 |
GO:0034389 |
lipid particle organization(GO:0034389) |
| 0.0 |
0.3 |
GO:0045040 |
protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 |
0.4 |
GO:0007368 |
determination of left/right symmetry(GO:0007368) specification of symmetry(GO:0009799) determination of bilateral symmetry(GO:0009855) |
| 0.0 |
0.1 |
GO:0043985 |
histone H4-R3 methylation(GO:0043985) |
| 0.0 |
0.2 |
GO:0006561 |
proline biosynthetic process(GO:0006561) |
| 0.0 |
0.1 |
GO:0006556 |
S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 |
0.3 |
GO:0043568 |
positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 |
0.6 |
GO:0032981 |
NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 |
0.1 |
GO:2000095 |
regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.0 |
1.2 |
GO:0071277 |
cellular response to calcium ion(GO:0071277) |
| 0.0 |
0.1 |
GO:0000491 |
small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.0 |
0.1 |
GO:0051152 |
positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 |
0.2 |
GO:0016255 |
attachment of GPI anchor to protein(GO:0016255) |
| 0.0 |
0.1 |
GO:1902626 |
assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 |
0.2 |
GO:0006544 |
glycine metabolic process(GO:0006544) |
| 0.0 |
0.3 |
GO:0006000 |
fructose metabolic process(GO:0006000) |
| 0.0 |
0.5 |
GO:0031297 |
replication fork processing(GO:0031297) |
| 0.0 |
0.4 |
GO:0001913 |
T cell mediated cytotoxicity(GO:0001913) |
| 0.0 |
0.2 |
GO:0019227 |
neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 |
0.6 |
GO:0006730 |
one-carbon metabolic process(GO:0006730) |
| 0.0 |
0.3 |
GO:1900383 |
regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 |
0.2 |
GO:0001867 |
complement activation, lectin pathway(GO:0001867) |
| 0.0 |
0.2 |
GO:0042126 |
nitrate metabolic process(GO:0042126) |
| 0.0 |
0.5 |
GO:2000811 |
negative regulation of anoikis(GO:2000811) |
| 0.0 |
0.3 |
GO:0035269 |
protein O-linked mannosylation(GO:0035269) |
| 0.0 |
0.9 |
GO:0033014 |
porphyrin-containing compound biosynthetic process(GO:0006779) tetrapyrrole biosynthetic process(GO:0033014) |
| 0.0 |
0.2 |
GO:0050858 |
negative regulation of antigen receptor-mediated signaling pathway(GO:0050858) negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 |
0.1 |
GO:0019287 |
isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.0 |
0.4 |
GO:0006767 |
water-soluble vitamin metabolic process(GO:0006767) |
| 0.0 |
0.1 |
GO:0001980 |
regulation of systemic arterial blood pressure by ischemic conditions(GO:0001980) |
| 0.0 |
0.1 |
GO:0006621 |
protein retention in ER lumen(GO:0006621) |
| 0.0 |
0.2 |
GO:2001014 |
regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.0 |
0.3 |
GO:0051894 |
positive regulation of focal adhesion assembly(GO:0051894) positive regulation of cell junction assembly(GO:1901890) |
| 0.0 |
0.8 |
GO:0030317 |
sperm motility(GO:0030317) |
| 0.0 |
0.1 |
GO:0010793 |
regulation of mRNA export from nucleus(GO:0010793) regulation of ribonucleoprotein complex localization(GO:2000197) |
| 0.0 |
0.6 |
GO:0035690 |
cellular response to drug(GO:0035690) |
| 0.0 |
0.1 |
GO:0015879 |
carnitine transport(GO:0015879) |
| 0.0 |
0.5 |
GO:0050435 |
beta-amyloid metabolic process(GO:0050435) |
| 0.0 |
1.5 |
GO:0051297 |
centrosome organization(GO:0051297) |
| 0.0 |
0.7 |
GO:0030819 |
positive regulation of cAMP biosynthetic process(GO:0030819) |
| 0.0 |
0.4 |
GO:0046325 |
negative regulation of glucose import(GO:0046325) |
| 0.0 |
0.3 |
GO:0045063 |
T-helper 1 cell differentiation(GO:0045063) |
| 0.0 |
0.4 |
GO:0095500 |
acetylcholine receptor signaling pathway(GO:0095500) postsynaptic signal transduction(GO:0098926) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.0 |
0.1 |
GO:0002523 |
leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.0 |
0.2 |
GO:0044243 |
collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 |
0.3 |
GO:0045747 |
positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 |
0.1 |
GO:0051168 |
nuclear export(GO:0051168) |
| 0.0 |
0.2 |
GO:0006491 |
N-glycan processing(GO:0006491) |
| 0.0 |
0.1 |
GO:0048305 |
immunoglobulin secretion(GO:0048305) |
| 0.0 |
0.5 |
GO:0032436 |
positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.0 |
0.2 |
GO:0036010 |
protein localization to endosome(GO:0036010) |
| 0.0 |
1.0 |
GO:0000079 |
regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) |
| 0.0 |
0.1 |
GO:0098532 |
histone H3-K27 trimethylation(GO:0098532) |
| 0.0 |
0.2 |
GO:0048172 |
regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 |
0.2 |
GO:0071260 |
cellular response to mechanical stimulus(GO:0071260) |
| 0.0 |
0.1 |
GO:1903069 |
regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903069) |
| 0.0 |
1.0 |
GO:0042147 |
retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 |
0.1 |
GO:0006102 |
isocitrate metabolic process(GO:0006102) |
| 0.0 |
0.2 |
GO:0097120 |
receptor localization to synapse(GO:0097120) |
| 0.0 |
0.1 |
GO:0006020 |
inositol metabolic process(GO:0006020) |
| 0.0 |
0.1 |
GO:0007019 |
microtubule depolymerization(GO:0007019) |
| 0.0 |
0.7 |
GO:0051453 |
regulation of intracellular pH(GO:0051453) |
| 0.0 |
0.3 |
GO:0046329 |
negative regulation of JNK cascade(GO:0046329) |
| 0.0 |
0.6 |
GO:0051289 |
protein homotetramerization(GO:0051289) |
| 0.0 |
0.1 |
GO:0001561 |
fatty acid alpha-oxidation(GO:0001561) |
| 0.0 |
0.5 |
GO:0010508 |
positive regulation of autophagy(GO:0010508) |
| 0.0 |
0.1 |
GO:0044253 |
positive regulation of collagen metabolic process(GO:0010714) positive regulation of collagen biosynthetic process(GO:0032967) positive regulation of multicellular organismal metabolic process(GO:0044253) |
| 0.0 |
0.5 |
GO:2001022 |
positive regulation of response to DNA damage stimulus(GO:2001022) |
| 0.0 |
0.2 |
GO:0072332 |
intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |