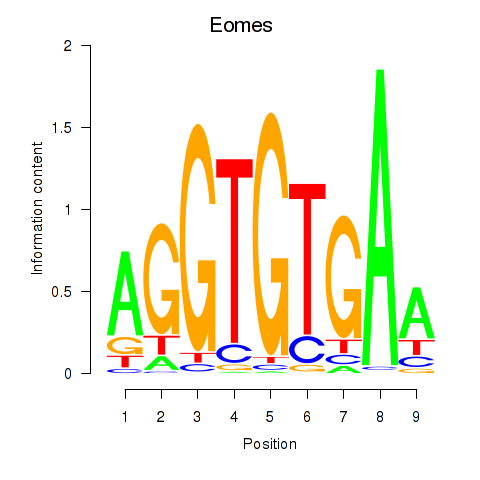
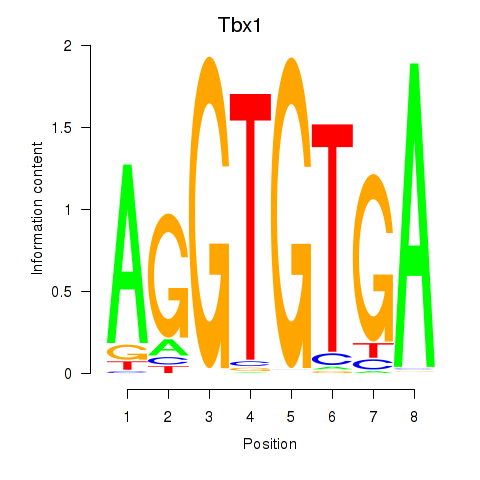
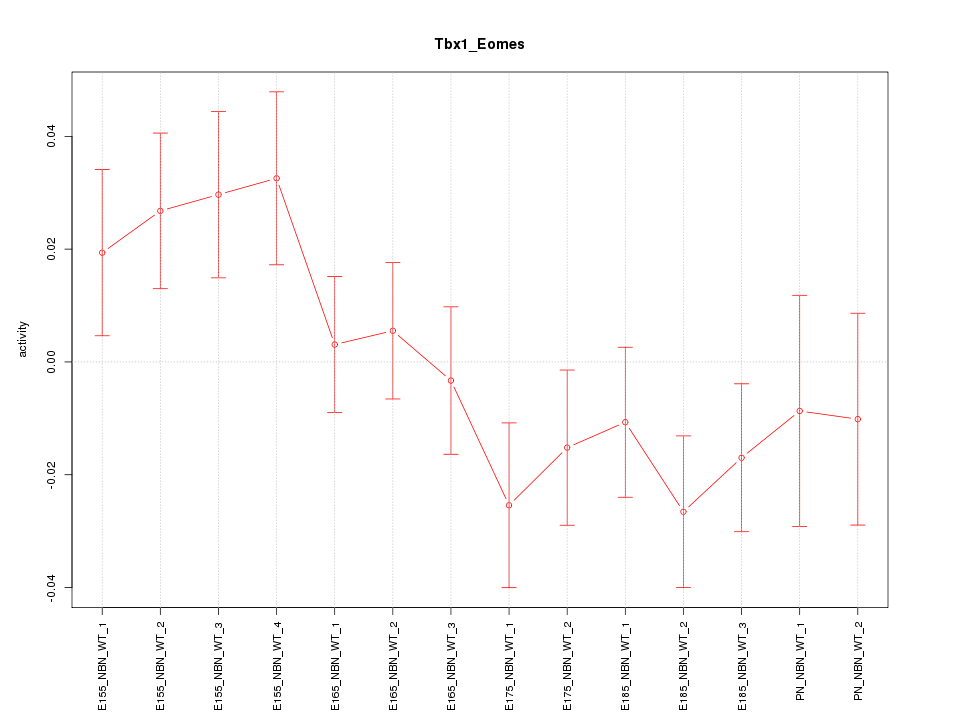
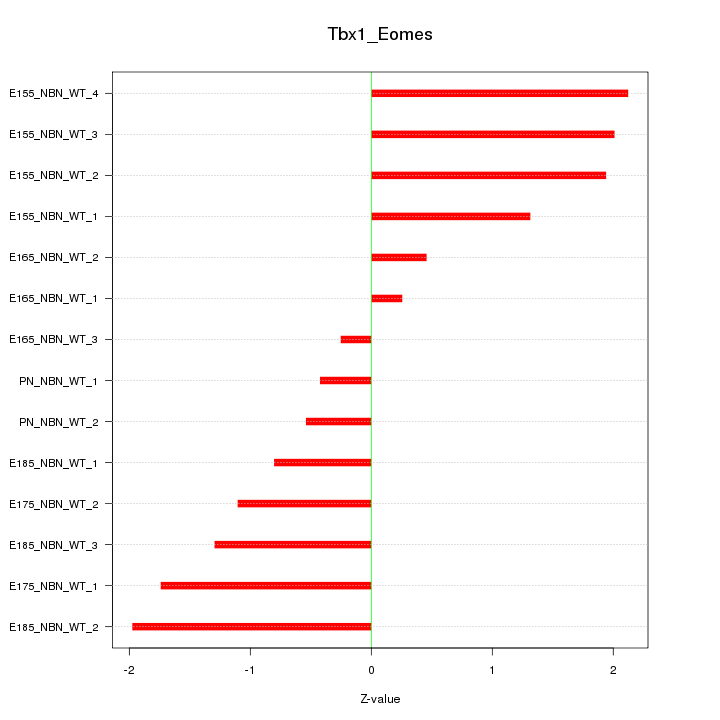
| 2.4 |
9.6 |
GO:0051866 |
general adaptation syndrome(GO:0051866) |
| 2.0 |
22.3 |
GO:0051967 |
negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 1.6 |
19.3 |
GO:0060159 |
regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.9 |
2.8 |
GO:0007621 |
negative regulation of female receptivity(GO:0007621) |
| 0.8 |
2.4 |
GO:0042663 |
regulation of endodermal cell fate specification(GO:0042663) |
| 0.8 |
2.3 |
GO:0070973 |
protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.6 |
4.0 |
GO:0034047 |
regulation of protein phosphatase type 2A activity(GO:0034047) |
| 0.5 |
1.6 |
GO:0003219 |
cardiac right ventricle formation(GO:0003219) |
| 0.5 |
5.9 |
GO:0060501 |
positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.5 |
1.4 |
GO:0007386 |
compartment pattern specification(GO:0007386) |
| 0.4 |
1.3 |
GO:0014005 |
microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.4 |
1.2 |
GO:0015772 |
disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.3 |
1.3 |
GO:0035616 |
histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.3 |
1.2 |
GO:2000795 |
negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.3 |
1.7 |
GO:0046549 |
retinal cone cell development(GO:0046549) |
| 0.3 |
1.0 |
GO:0015871 |
choline transport(GO:0015871) |
| 0.2 |
6.8 |
GO:0035335 |
peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.2 |
10.3 |
GO:0001919 |
regulation of receptor recycling(GO:0001919) |
| 0.2 |
0.6 |
GO:0036118 |
hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.2 |
0.6 |
GO:0090526 |
response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.2 |
1.0 |
GO:0002158 |
osteoclast proliferation(GO:0002158) |
| 0.2 |
0.5 |
GO:0006546 |
glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.2 |
0.9 |
GO:1903847 |
regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 0.2 |
0.8 |
GO:0007171 |
cardiac conduction system development(GO:0003161) activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.1 |
0.5 |
GO:0061030 |
axon target recognition(GO:0007412) epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.1 |
0.9 |
GO:1900194 |
negative regulation of oocyte maturation(GO:1900194) |
| 0.1 |
1.3 |
GO:0097151 |
positive regulation of inhibitory postsynaptic potential(GO:0097151) |
| 0.1 |
0.5 |
GO:0038128 |
ERBB2 signaling pathway(GO:0038128) |
| 0.1 |
1.2 |
GO:0019227 |
neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 |
1.1 |
GO:0060059 |
embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.1 |
0.5 |
GO:0035262 |
gonad morphogenesis(GO:0035262) |
| 0.1 |
1.6 |
GO:0003334 |
keratinocyte development(GO:0003334) |
| 0.1 |
0.3 |
GO:0050915 |
sensory perception of sour taste(GO:0050915) |
| 0.1 |
0.2 |
GO:0097623 |
potassium ion export across plasma membrane(GO:0097623) |
| 0.1 |
0.3 |
GO:0050913 |
sensory perception of bitter taste(GO:0050913) |
| 0.1 |
1.9 |
GO:0042149 |
cellular response to glucose starvation(GO:0042149) |
| 0.1 |
0.6 |
GO:0010457 |
centriole-centriole cohesion(GO:0010457) |
| 0.1 |
0.6 |
GO:2001256 |
regulation of store-operated calcium entry(GO:2001256) |
| 0.1 |
0.4 |
GO:0071285 |
cellular response to lithium ion(GO:0071285) |
| 0.1 |
0.2 |
GO:0035356 |
cellular triglyceride homeostasis(GO:0035356) |
| 0.0 |
0.8 |
GO:0099500 |
synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.0 |
2.6 |
GO:0008347 |
glial cell migration(GO:0008347) |
| 0.0 |
0.4 |
GO:0000185 |
activation of MAPKKK activity(GO:0000185) |
| 0.0 |
0.5 |
GO:0060481 |
lobar bronchus epithelium development(GO:0060481) |
| 0.0 |
2.0 |
GO:0060612 |
adipose tissue development(GO:0060612) |
| 0.0 |
6.2 |
GO:0050807 |
regulation of synapse organization(GO:0050807) |
| 0.0 |
0.3 |
GO:0036506 |
maintenance of unfolded protein(GO:0036506) protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.0 |
0.3 |
GO:0030035 |
microspike assembly(GO:0030035) |
| 0.0 |
0.4 |
GO:0032876 |
negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 |
0.7 |
GO:0010842 |
retina layer formation(GO:0010842) |
| 0.0 |
0.2 |
GO:2000676 |
positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.0 |
0.1 |
GO:1900365 |
positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.0 |
0.4 |
GO:0006120 |
mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 |
0.0 |
GO:0060523 |
prostate epithelial cord elongation(GO:0060523) |
| 0.0 |
0.2 |
GO:2000741 |
positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.0 |
0.1 |
GO:0034729 |
histone H3-K79 methylation(GO:0034729) |
| 0.0 |
0.4 |
GO:0042773 |
ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 |
0.3 |
GO:0030953 |
astral microtubule organization(GO:0030953) |
| 0.0 |
0.2 |
GO:0006614 |
SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 |
0.3 |
GO:0019221 |
cytokine-mediated signaling pathway(GO:0019221) |
| 0.0 |
0.3 |
GO:0032967 |
positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.0 |
0.1 |
GO:0034312 |
diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) sphingoid biosynthetic process(GO:0046520) |
| 0.0 |
0.7 |
GO:0030500 |
regulation of bone mineralization(GO:0030500) |
| 0.0 |
0.2 |
GO:0048387 |
negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 |
0.1 |
GO:0017196 |
N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 |
0.0 |
GO:0035521 |
monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.0 |
0.5 |
GO:0046513 |
ceramide biosynthetic process(GO:0046513) |