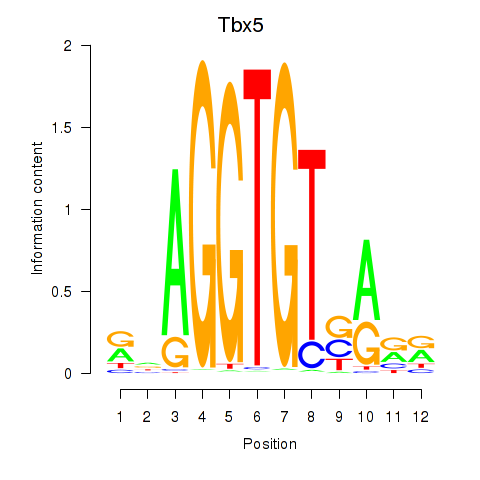
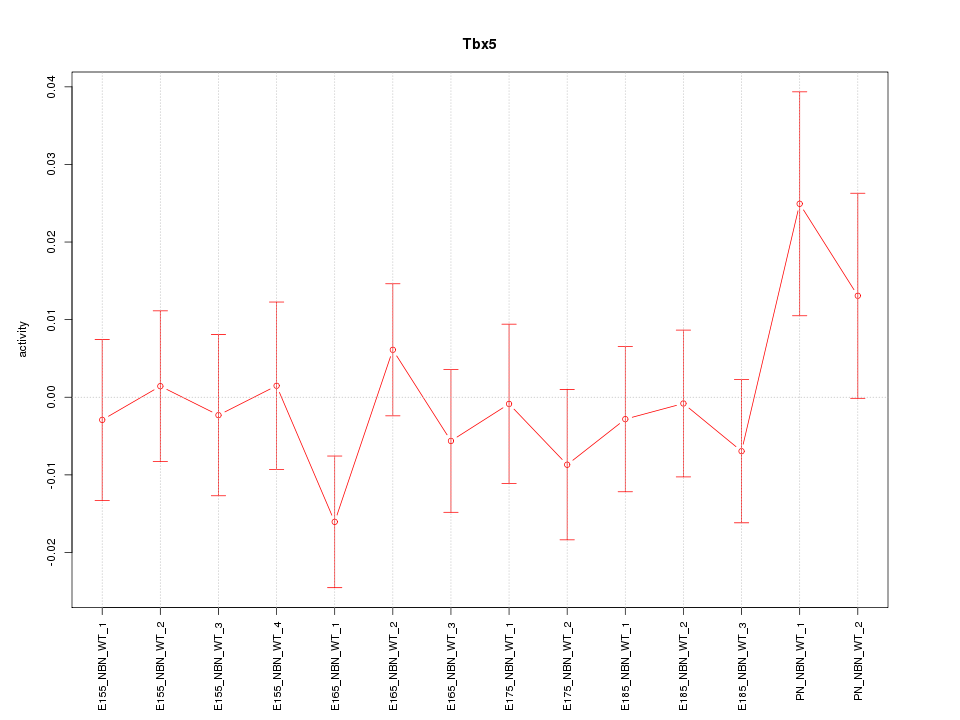
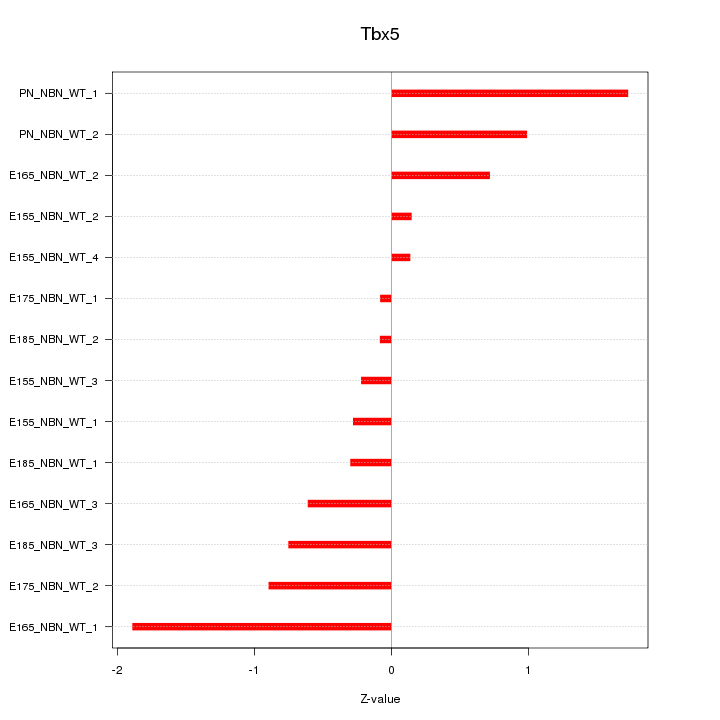
| 0.4 |
1.3 |
GO:0021529 |
spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.4 |
2.1 |
GO:0035022 |
positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.3 |
1.0 |
GO:0003245 |
cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.3 |
2.7 |
GO:0016338 |
calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.3 |
1.5 |
GO:0072488 |
ammonium transmembrane transport(GO:0072488) |
| 0.3 |
0.8 |
GO:0007521 |
muscle cell fate determination(GO:0007521) mammary placode formation(GO:0060596) |
| 0.3 |
0.3 |
GO:0003241 |
growth involved in heart morphogenesis(GO:0003241) |
| 0.2 |
0.7 |
GO:0007210 |
serotonin receptor signaling pathway(GO:0007210) |
| 0.2 |
0.6 |
GO:0034162 |
toll-like receptor 9 signaling pathway(GO:0034162) |
| 0.2 |
1.2 |
GO:0060666 |
dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.2 |
0.8 |
GO:0007386 |
compartment pattern specification(GO:0007386) |
| 0.2 |
0.8 |
GO:0051365 |
cellular response to potassium ion starvation(GO:0051365) |
| 0.1 |
0.7 |
GO:0010025 |
wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.1 |
0.5 |
GO:1902608 |
regulation of large conductance calcium-activated potassium channel activity(GO:1902606) positive regulation of large conductance calcium-activated potassium channel activity(GO:1902608) |
| 0.1 |
0.6 |
GO:0030091 |
protein repair(GO:0030091) |
| 0.1 |
0.5 |
GO:1900145 |
regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900175) positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.1 |
0.7 |
GO:0043415 |
positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.1 |
0.3 |
GO:0090327 |
negative regulation of locomotion involved in locomotory behavior(GO:0090327) |
| 0.1 |
0.3 |
GO:0003349 |
epicardium-derived cardiac endothelial cell differentiation(GO:0003349) |
| 0.1 |
1.1 |
GO:0090527 |
actin filament reorganization(GO:0090527) |
| 0.1 |
0.4 |
GO:0003192 |
mitral valve formation(GO:0003192) cell migration involved in endocardial cushion formation(GO:0003273) condensed mesenchymal cell proliferation(GO:0072137) |
| 0.1 |
1.0 |
GO:0007168 |
receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.1 |
1.2 |
GO:0060272 |
embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.1 |
0.4 |
GO:0046013 |
regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.1 |
0.9 |
GO:0090385 |
phagosome-lysosome fusion(GO:0090385) |
| 0.1 |
0.5 |
GO:0050861 |
positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.1 |
0.2 |
GO:2001187 |
positive regulation of CD8-positive, alpha-beta T cell activation(GO:2001187) |
| 0.1 |
0.8 |
GO:0019227 |
neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 |
0.3 |
GO:0071139 |
positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) resolution of recombination intermediates(GO:0071139) |
| 0.1 |
0.5 |
GO:0000022 |
mitotic spindle elongation(GO:0000022) mitotic spindle midzone assembly(GO:0051256) |
| 0.1 |
0.5 |
GO:0007342 |
fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.1 |
0.2 |
GO:0001951 |
intestinal D-glucose absorption(GO:0001951) terminal web assembly(GO:1902896) |
| 0.1 |
0.2 |
GO:0042138 |
meiotic DNA double-strand break formation(GO:0042138) |
| 0.1 |
0.4 |
GO:1903352 |
ornithine transport(GO:0015822) L-ornithine transmembrane transport(GO:1903352) |
| 0.1 |
0.3 |
GO:0010046 |
arginine biosynthetic process(GO:0006526) response to mycotoxin(GO:0010046) |
| 0.1 |
3.3 |
GO:0043666 |
regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.1 |
0.2 |
GO:0030214 |
hyaluronan catabolic process(GO:0030214) |
| 0.1 |
0.4 |
GO:0030382 |
sperm mitochondrion organization(GO:0030382) |
| 0.1 |
0.5 |
GO:0010587 |
miRNA catabolic process(GO:0010587) |
| 0.1 |
0.2 |
GO:0042276 |
error-prone translesion synthesis(GO:0042276) |
| 0.1 |
0.4 |
GO:0090195 |
chemokine secretion(GO:0090195) regulation of chemokine secretion(GO:0090196) positive regulation of chemokine secretion(GO:0090197) |
| 0.1 |
0.2 |
GO:0009812 |
flavonoid metabolic process(GO:0009812) flavonoid biosynthetic process(GO:0009813) flavonoid glucuronidation(GO:0052696) |
| 0.1 |
0.7 |
GO:0016264 |
gap junction assembly(GO:0016264) |
| 0.1 |
0.4 |
GO:0038028 |
insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.1 |
0.5 |
GO:0006691 |
leukotriene metabolic process(GO:0006691) |
| 0.1 |
0.9 |
GO:0009109 |
coenzyme catabolic process(GO:0009109) |
| 0.1 |
0.3 |
GO:0036006 |
response to macrophage colony-stimulating factor(GO:0036005) cellular response to macrophage colony-stimulating factor stimulus(GO:0036006) |
| 0.1 |
0.4 |
GO:0043383 |
negative T cell selection(GO:0043383) negative thymic T cell selection(GO:0045060) |
| 0.1 |
1.0 |
GO:0051150 |
regulation of smooth muscle cell differentiation(GO:0051150) |
| 0.1 |
0.2 |
GO:0097278 |
complement-dependent cytotoxicity(GO:0097278) |
| 0.1 |
0.2 |
GO:0009446 |
putrescine biosynthetic process(GO:0009446) |
| 0.1 |
0.4 |
GO:0009449 |
gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.1 |
0.2 |
GO:1904451 |
regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904451) positive regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904453) |
| 0.0 |
0.8 |
GO:0006953 |
acute-phase response(GO:0006953) |
| 0.0 |
0.2 |
GO:0001575 |
globoside metabolic process(GO:0001575) |
| 0.0 |
0.4 |
GO:0036158 |
outer dynein arm assembly(GO:0036158) |
| 0.0 |
0.5 |
GO:0045647 |
negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 |
0.2 |
GO:0015871 |
choline transport(GO:0015871) |
| 0.0 |
0.1 |
GO:0060385 |
axonogenesis involved in innervation(GO:0060385) |
| 0.0 |
0.4 |
GO:0045974 |
miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 |
0.1 |
GO:1903031 |
regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.0 |
0.4 |
GO:0010457 |
centriole-centriole cohesion(GO:0010457) |
| 0.0 |
0.3 |
GO:0031053 |
primary miRNA processing(GO:0031053) |
| 0.0 |
0.1 |
GO:0030167 |
proteoglycan catabolic process(GO:0030167) |
| 0.0 |
0.2 |
GO:0016576 |
histone dephosphorylation(GO:0016576) |
| 0.0 |
0.1 |
GO:0097167 |
circadian regulation of translation(GO:0097167) |
| 0.0 |
0.2 |
GO:1900451 |
positive regulation of glutamate receptor signaling pathway(GO:1900451) positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 0.0 |
0.2 |
GO:1903753 |
cortical microtubule organization(GO:0043622) establishment of centrosome localization(GO:0051660) negative regulation of p38MAPK cascade(GO:1903753) |
| 0.0 |
0.2 |
GO:0045162 |
clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 |
0.5 |
GO:0000082 |
G1/S transition of mitotic cell cycle(GO:0000082) |
| 0.0 |
0.1 |
GO:0035624 |
receptor transactivation(GO:0035624) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) |
| 0.0 |
0.3 |
GO:0036506 |
maintenance of unfolded protein(GO:0036506) protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.0 |
0.6 |
GO:0035024 |
negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 |
0.3 |
GO:0019368 |
fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 |
0.1 |
GO:1900157 |
regulation of bone mineralization involved in bone maturation(GO:1900157) |
| 0.0 |
0.1 |
GO:1903587 |
regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) |
| 0.0 |
0.5 |
GO:0006903 |
vesicle targeting(GO:0006903) |
| 0.0 |
0.8 |
GO:0003333 |
amino acid transmembrane transport(GO:0003333) |
| 0.0 |
0.1 |
GO:0035262 |
gonad morphogenesis(GO:0035262) |
| 0.0 |
0.2 |
GO:0031508 |
pericentric heterochromatin assembly(GO:0031508) |
| 0.0 |
1.4 |
GO:0015909 |
long-chain fatty acid transport(GO:0015909) |
| 0.0 |
0.1 |
GO:0071544 |
diphosphoinositol polyphosphate metabolic process(GO:0071543) diphosphoinositol polyphosphate catabolic process(GO:0071544) |
| 0.0 |
0.2 |
GO:0006621 |
protein retention in ER lumen(GO:0006621) |
| 0.0 |
0.4 |
GO:0072010 |
renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.0 |
0.7 |
GO:0071108 |
protein K48-linked deubiquitination(GO:0071108) |
| 0.0 |
0.2 |
GO:0030854 |
positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 |
0.3 |
GO:0043589 |
skin morphogenesis(GO:0043589) |
| 0.0 |
0.2 |
GO:0009992 |
cellular water homeostasis(GO:0009992) |
| 0.0 |
0.1 |
GO:2000501 |
natural killer cell chemotaxis(GO:0035747) regulation of natural killer cell chemotaxis(GO:2000501) |
| 0.0 |
0.2 |
GO:0021780 |
oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.0 |
0.7 |
GO:0048791 |
calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 |
0.4 |
GO:0006958 |
complement activation, classical pathway(GO:0006958) |
| 0.0 |
0.4 |
GO:0019915 |
lipid storage(GO:0019915) |
| 0.0 |
0.4 |
GO:0060236 |
regulation of mitotic spindle organization(GO:0060236) |
| 0.0 |
0.8 |
GO:0001919 |
regulation of receptor recycling(GO:0001919) |
| 0.0 |
0.7 |
GO:0000266 |
mitochondrial fission(GO:0000266) |
| 0.0 |
0.0 |
GO:0010744 |
positive regulation of macrophage derived foam cell differentiation(GO:0010744) positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.0 |
0.2 |
GO:0007130 |
synaptonemal complex assembly(GO:0007130) |
| 0.0 |
0.4 |
GO:0006471 |
protein ADP-ribosylation(GO:0006471) |
| 0.0 |
0.7 |
GO:0045103 |
intermediate filament-based process(GO:0045103) |
| 0.0 |
0.2 |
GO:0048702 |
embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 |
0.1 |
GO:0016255 |
attachment of GPI anchor to protein(GO:0016255) |
| 0.0 |
0.2 |
GO:0045792 |
negative regulation of cell size(GO:0045792) |
| 0.0 |
0.2 |
GO:0048146 |
positive regulation of fibroblast proliferation(GO:0048146) |
| 0.0 |
0.7 |
GO:0031114 |
negative regulation of microtubule depolymerization(GO:0007026) regulation of microtubule depolymerization(GO:0031114) |
| 0.0 |
0.2 |
GO:0007250 |
activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 |
0.5 |
GO:0070830 |
bicellular tight junction assembly(GO:0070830) |
| 0.0 |
0.1 |
GO:0016584 |
nucleosome positioning(GO:0016584) |
| 0.0 |
0.1 |
GO:1901660 |
calcium ion export(GO:1901660) |
| 0.0 |
0.1 |
GO:0032911 |
nerve growth factor production(GO:0032902) negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.0 |
0.4 |
GO:0035335 |
peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 |
0.4 |
GO:0045604 |
regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 |
0.6 |
GO:0051893 |
regulation of focal adhesion assembly(GO:0051893) regulation of cell-substrate junction assembly(GO:0090109) |
| 0.0 |
0.1 |
GO:2001022 |
positive regulation of response to DNA damage stimulus(GO:2001022) |
| 0.0 |
0.3 |
GO:0000038 |
very long-chain fatty acid metabolic process(GO:0000038) |
| 0.0 |
0.2 |
GO:0032463 |
negative regulation of protein homooligomerization(GO:0032463) |
| 0.0 |
0.5 |
GO:0033119 |
negative regulation of RNA splicing(GO:0033119) |
| 0.0 |
0.2 |
GO:0048387 |
negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 |
0.1 |
GO:0032367 |
intracellular cholesterol transport(GO:0032367) |
| 0.0 |
0.3 |
GO:1900026 |
positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 |
0.1 |
GO:0035269 |
protein O-linked mannosylation(GO:0035269) |
| 0.0 |
0.1 |
GO:0010818 |
T cell chemotaxis(GO:0010818) |