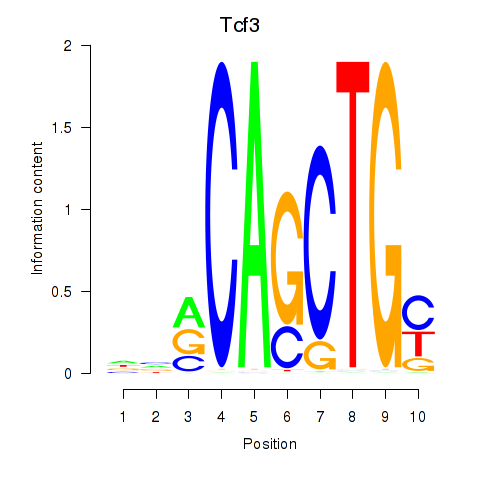
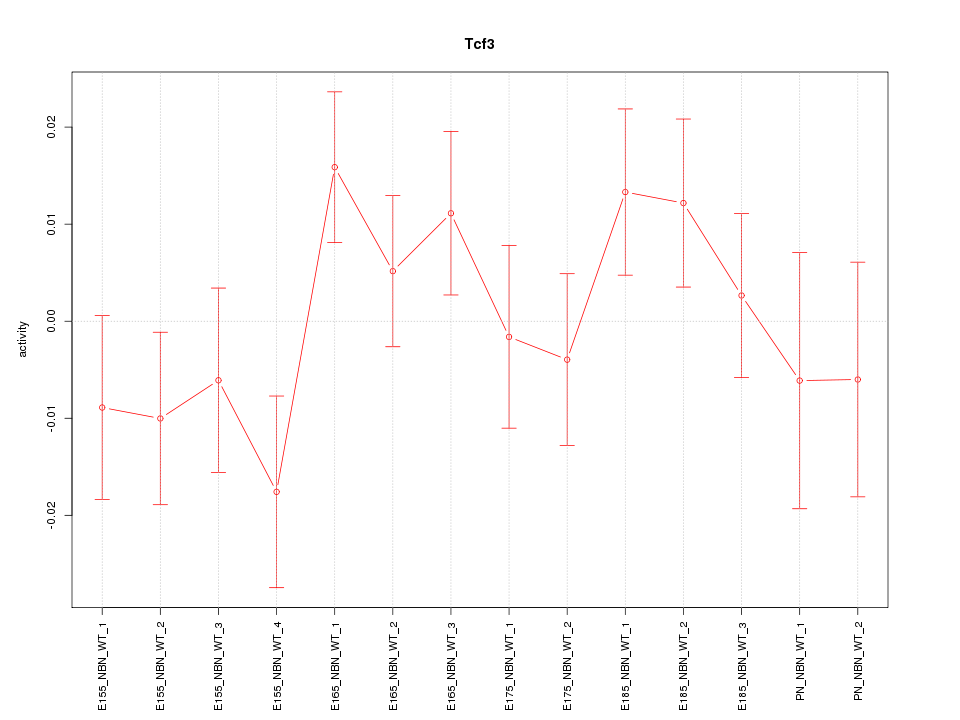
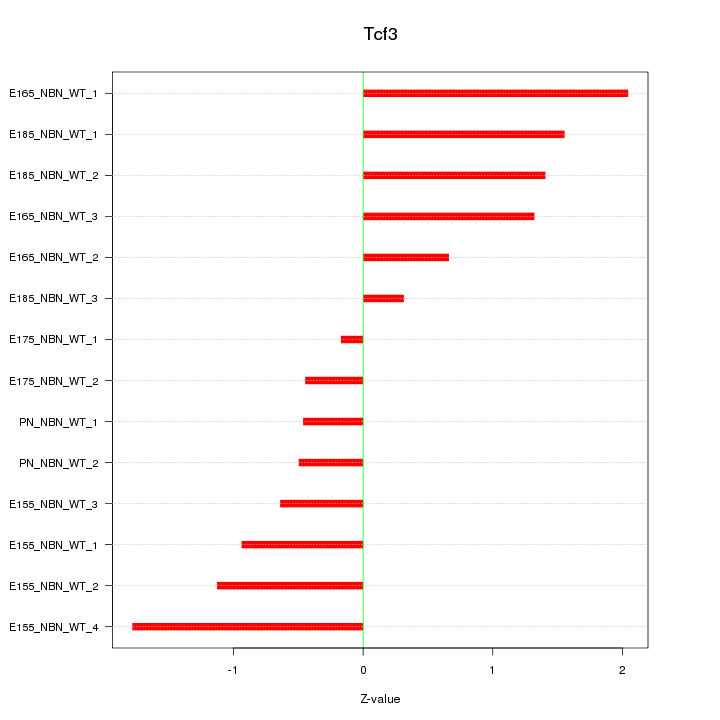
| 0.8 |
2.4 |
GO:0032765 |
positive regulation of mast cell cytokine production(GO:0032765) |
| 0.6 |
1.8 |
GO:0006601 |
creatine biosynthetic process(GO:0006601) |
| 0.5 |
1.5 |
GO:0021649 |
vestibulocochlear nerve morphogenesis(GO:0021648) vestibulocochlear nerve structural organization(GO:0021649) ganglion morphogenesis(GO:0061552) dorsal root ganglion morphogenesis(GO:1904835) |
| 0.4 |
1.6 |
GO:0051725 |
protein de-ADP-ribosylation(GO:0051725) |
| 0.4 |
2.2 |
GO:0050861 |
positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.4 |
1.1 |
GO:0010752 |
signal complex assembly(GO:0007172) regulation of cGMP-mediated signaling(GO:0010752) |
| 0.3 |
1.6 |
GO:0048133 |
germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.3 |
0.8 |
GO:2000507 |
positive regulation of energy homeostasis(GO:2000507) |
| 0.2 |
0.7 |
GO:0021837 |
motogenic signaling involved in postnatal olfactory bulb interneuron migration(GO:0021837) positive regulation of mitotic cell cycle DNA replication(GO:1903465) |
| 0.2 |
0.7 |
GO:0044333 |
Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.2 |
0.6 |
GO:0042939 |
renal sodium ion transport(GO:0003096) glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.2 |
0.8 |
GO:0072235 |
distal convoluted tubule development(GO:0072025) DCT cell differentiation(GO:0072069) metanephric distal convoluted tubule development(GO:0072221) metanephric distal tubule development(GO:0072235) metanephric DCT cell differentiation(GO:0072240) |
| 0.2 |
0.4 |
GO:1990791 |
dorsal root ganglion development(GO:1990791) |
| 0.2 |
2.6 |
GO:0097067 |
negative regulation of erythrocyte differentiation(GO:0045647) cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.2 |
1.9 |
GO:0034983 |
peptidyl-lysine deacetylation(GO:0034983) regulation of skeletal muscle fiber development(GO:0048742) |
| 0.2 |
0.4 |
GO:0002159 |
desmosome assembly(GO:0002159) |
| 0.2 |
0.9 |
GO:0070327 |
thyroid hormone transport(GO:0070327) |
| 0.2 |
0.4 |
GO:0072289 |
metanephric nephron tubule formation(GO:0072289) |
| 0.2 |
0.9 |
GO:0032224 |
positive regulation of synaptic transmission, cholinergic(GO:0032224) |
| 0.2 |
0.9 |
GO:0002901 |
mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.2 |
0.5 |
GO:1900673 |
olefin metabolic process(GO:1900673) |
| 0.2 |
1.0 |
GO:0060666 |
dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.2 |
0.5 |
GO:0050855 |
regulation of B cell receptor signaling pathway(GO:0050855) |
| 0.2 |
0.5 |
GO:0098528 |
terpenoid catabolic process(GO:0016115) skeletal muscle fiber differentiation(GO:0098528) |
| 0.1 |
0.4 |
GO:0032650 |
regulation of interleukin-1 alpha production(GO:0032650) positive regulation of interleukin-1 alpha production(GO:0032730) cardiac cell fate determination(GO:0060913) |
| 0.1 |
1.0 |
GO:0009449 |
gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.1 |
0.6 |
GO:0048539 |
bone marrow development(GO:0048539) |
| 0.1 |
0.4 |
GO:0034162 |
toll-like receptor 9 signaling pathway(GO:0034162) |
| 0.1 |
0.9 |
GO:0031666 |
positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.1 |
0.5 |
GO:1900740 |
regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.1 |
0.9 |
GO:0060923 |
cardiac muscle cell fate commitment(GO:0060923) |
| 0.1 |
1.0 |
GO:0000050 |
urea cycle(GO:0000050) urea metabolic process(GO:0019627) |
| 0.1 |
0.8 |
GO:2000491 |
positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.1 |
0.5 |
GO:0016340 |
calcium-dependent cell-matrix adhesion(GO:0016340) |
| 0.1 |
1.5 |
GO:0021527 |
spinal cord association neuron differentiation(GO:0021527) |
| 0.1 |
0.3 |
GO:0045014 |
detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) detection of glucose(GO:0051594) |
| 0.1 |
0.7 |
GO:0032534 |
regulation of microvillus assembly(GO:0032534) |
| 0.1 |
0.7 |
GO:0007342 |
fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.1 |
0.4 |
GO:1903587 |
regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) |
| 0.1 |
0.7 |
GO:0010820 |
positive regulation of T cell chemotaxis(GO:0010820) |
| 0.1 |
0.5 |
GO:2000680 |
regulation of rubidium ion transport(GO:2000680) |
| 0.1 |
0.4 |
GO:0060715 |
syncytiotrophoblast cell differentiation involved in labyrinthine layer development(GO:0060715) |
| 0.1 |
0.5 |
GO:0010668 |
ectodermal cell differentiation(GO:0010668) |
| 0.1 |
0.3 |
GO:0003100 |
regulation of systemic arterial blood pressure by endothelin(GO:0003100) cellular magnesium ion homeostasis(GO:0010961) vein smooth muscle contraction(GO:0014826) |
| 0.1 |
0.4 |
GO:0010288 |
response to lead ion(GO:0010288) |
| 0.1 |
2.0 |
GO:0010614 |
negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.1 |
0.3 |
GO:1903048 |
regulation of acetylcholine-gated cation channel activity(GO:1903048) |
| 0.1 |
0.5 |
GO:0034227 |
tRNA thio-modification(GO:0034227) |
| 0.1 |
0.3 |
GO:0003032 |
detection of oxygen(GO:0003032) regulation of epithelial to mesenchymal transition involved in endocardial cushion formation(GO:1905005) |
| 0.1 |
0.7 |
GO:0021678 |
third ventricle development(GO:0021678) |
| 0.1 |
0.3 |
GO:0045578 |
negative regulation of B cell differentiation(GO:0045578) |
| 0.1 |
0.4 |
GO:0071500 |
cellular response to nitrosative stress(GO:0071500) |
| 0.1 |
0.8 |
GO:0009404 |
toxin metabolic process(GO:0009404) |
| 0.1 |
0.5 |
GO:2001054 |
negative regulation of mesenchymal cell apoptotic process(GO:2001054) |
| 0.1 |
0.3 |
GO:1990414 |
replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.1 |
0.3 |
GO:0010637 |
negative regulation of mitochondrial fusion(GO:0010637) |
| 0.1 |
0.1 |
GO:1901420 |
negative regulation of response to alcohol(GO:1901420) |
| 0.1 |
0.2 |
GO:0015938 |
coenzyme A catabolic process(GO:0015938) nucleoside bisphosphate catabolic process(GO:0033869) ribonucleoside bisphosphate catabolic process(GO:0034031) purine nucleoside bisphosphate catabolic process(GO:0034034) |
| 0.1 |
0.2 |
GO:2000409 |
positive regulation of T cell extravasation(GO:2000409) |
| 0.1 |
0.3 |
GO:0072610 |
myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) interleukin-12 secretion(GO:0072610) regulation of interleukin-12 secretion(GO:2001182) positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.1 |
0.4 |
GO:0015862 |
uridine transport(GO:0015862) |
| 0.1 |
0.2 |
GO:0086017 |
Purkinje myocyte action potential(GO:0086017) |
| 0.1 |
0.2 |
GO:0046880 |
regulation of follicle-stimulating hormone secretion(GO:0046880) follicle-stimulating hormone secretion(GO:0046884) |
| 0.1 |
0.2 |
GO:1903288 |
regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) |
| 0.1 |
0.6 |
GO:0043252 |
sodium-independent organic anion transport(GO:0043252) |
| 0.1 |
0.4 |
GO:0045200 |
establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.1 |
0.8 |
GO:0033623 |
regulation of integrin activation(GO:0033623) |
| 0.1 |
0.2 |
GO:2001137 |
positive regulation of endocytic recycling(GO:2001137) |
| 0.1 |
0.2 |
GO:0046340 |
diacylglycerol catabolic process(GO:0046340) |
| 0.1 |
0.2 |
GO:0019074 |
viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.1 |
1.2 |
GO:0032967 |
positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.1 |
0.2 |
GO:0097623 |
potassium ion export across plasma membrane(GO:0097623) |
| 0.1 |
0.2 |
GO:0002949 |
tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.1 |
0.5 |
GO:2000194 |
regulation of female gonad development(GO:2000194) |
| 0.1 |
0.2 |
GO:0021612 |
facial nerve structural organization(GO:0021612) |
| 0.1 |
0.2 |
GO:0050925 |
negative regulation of negative chemotaxis(GO:0050925) |
| 0.0 |
0.8 |
GO:0044243 |
collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 |
0.3 |
GO:0005981 |
regulation of glycogen catabolic process(GO:0005981) |
| 0.0 |
0.6 |
GO:0000188 |
inactivation of MAPK activity(GO:0000188) |
| 0.0 |
0.2 |
GO:0046598 |
positive regulation of viral entry into host cell(GO:0046598) |
| 0.0 |
0.8 |
GO:0038166 |
angiotensin-activated signaling pathway(GO:0038166) |
| 0.0 |
0.2 |
GO:0010724 |
regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 |
1.0 |
GO:2000310 |
regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.0 |
0.2 |
GO:0006564 |
L-serine biosynthetic process(GO:0006564) |
| 0.0 |
0.1 |
GO:0035574 |
histone H4-K20 demethylation(GO:0035574) |
| 0.0 |
0.2 |
GO:0045657 |
positive regulation of monocyte differentiation(GO:0045657) |
| 0.0 |
0.2 |
GO:0033580 |
protein glycosylation at cell surface(GO:0033575) protein galactosylation at cell surface(GO:0033580) protein galactosylation(GO:0042125) |
| 0.0 |
0.1 |
GO:1902953 |
positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) |
| 0.0 |
0.1 |
GO:0015014 |
heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 |
0.2 |
GO:0007023 |
post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 |
0.3 |
GO:0015791 |
polyol transport(GO:0015791) |
| 0.0 |
0.2 |
GO:0008295 |
spermidine biosynthetic process(GO:0008295) |
| 0.0 |
0.1 |
GO:0035864 |
response to potassium ion(GO:0035864) cellular response to potassium ion(GO:0035865) |
| 0.0 |
0.2 |
GO:0032911 |
nerve growth factor production(GO:0032902) negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.0 |
0.2 |
GO:0007258 |
JUN phosphorylation(GO:0007258) |
| 0.0 |
1.1 |
GO:1901522 |
positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.0 |
1.3 |
GO:0007026 |
negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 |
0.2 |
GO:2000774 |
positive regulation of cellular senescence(GO:2000774) |
| 0.0 |
0.2 |
GO:0006172 |
ADP biosynthetic process(GO:0006172) |
| 0.0 |
0.3 |
GO:0001867 |
complement activation, lectin pathway(GO:0001867) |
| 0.0 |
0.2 |
GO:0051461 |
positive regulation of heat generation(GO:0031652) drinking behavior(GO:0042756) regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 0.0 |
0.3 |
GO:0005513 |
detection of calcium ion(GO:0005513) |
| 0.0 |
0.1 |
GO:0021577 |
hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.0 |
0.8 |
GO:0048268 |
clathrin coat assembly(GO:0048268) |
| 0.0 |
0.1 |
GO:0019509 |
L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 |
0.1 |
GO:0048739 |
cardiac muscle fiber development(GO:0048739) |
| 0.0 |
0.6 |
GO:0042182 |
ketone catabolic process(GO:0042182) |
| 0.0 |
0.3 |
GO:0090527 |
actin filament reorganization(GO:0090527) |
| 0.0 |
0.6 |
GO:0060080 |
inhibitory postsynaptic potential(GO:0060080) |
| 0.0 |
0.2 |
GO:1990564 |
protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 |
0.2 |
GO:0045059 |
positive thymic T cell selection(GO:0045059) |
| 0.0 |
0.3 |
GO:0015868 |
purine ribonucleotide transport(GO:0015868) |
| 0.0 |
0.1 |
GO:0097051 |
establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) |
| 0.0 |
0.4 |
GO:0006706 |
steroid catabolic process(GO:0006706) |
| 0.0 |
0.3 |
GO:0031547 |
brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 0.0 |
0.1 |
GO:1904690 |
adenosine to inosine editing(GO:0006382) regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 0.0 |
0.5 |
GO:0034142 |
toll-like receptor 4 signaling pathway(GO:0034142) |
| 0.0 |
0.4 |
GO:0002495 |
antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.0 |
0.2 |
GO:0042572 |
retinol metabolic process(GO:0042572) |
| 0.0 |
0.2 |
GO:0071803 |
positive regulation of podosome assembly(GO:0071803) |
| 0.0 |
0.5 |
GO:1900153 |
regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.0 |
0.2 |
GO:0051036 |
regulation of endosome size(GO:0051036) |
| 0.0 |
0.3 |
GO:0010748 |
negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.0 |
0.2 |
GO:0021942 |
radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.0 |
0.2 |
GO:0006450 |
regulation of translational fidelity(GO:0006450) |
| 0.0 |
0.2 |
GO:0030432 |
peristalsis(GO:0030432) |
| 0.0 |
0.1 |
GO:0009992 |
cellular water homeostasis(GO:0009992) |
| 0.0 |
0.3 |
GO:2000353 |
positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.0 |
0.5 |
GO:0040034 |
regulation of development, heterochronic(GO:0040034) |
| 0.0 |
0.5 |
GO:0050873 |
brown fat cell differentiation(GO:0050873) |
| 0.0 |
0.2 |
GO:1900262 |
regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 |
0.4 |
GO:0045662 |
negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 |
0.1 |
GO:2000210 |
positive regulation of anoikis(GO:2000210) |
| 0.0 |
0.1 |
GO:1901475 |
pyruvate transmembrane transport(GO:1901475) |
| 0.0 |
0.2 |
GO:0007175 |
negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 |
0.2 |
GO:1904587 |
glycoprotein ERAD pathway(GO:0097466) response to glycoprotein(GO:1904587) |
| 0.0 |
0.1 |
GO:0090038 |
negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 |
0.9 |
GO:0043242 |
negative regulation of protein complex disassembly(GO:0043242) negative regulation of protein depolymerization(GO:1901880) |
| 0.0 |
0.5 |
GO:0007205 |
protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 |
0.1 |
GO:2000832 |
protein-chromophore linkage(GO:0018298) negative regulation of steroid hormone secretion(GO:2000832) negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.0 |
0.3 |
GO:0071218 |
cellular response to misfolded protein(GO:0071218) |
| 0.0 |
0.1 |
GO:0060017 |
parathyroid gland development(GO:0060017) |
| 0.0 |
0.1 |
GO:0032513 |
negative regulation of protein phosphatase type 2B activity(GO:0032513) |
| 0.0 |
0.1 |
GO:0042534 |
tumor necrosis factor biosynthetic process(GO:0042533) regulation of tumor necrosis factor biosynthetic process(GO:0042534) |
| 0.0 |
0.6 |
GO:0034629 |
cellular protein complex localization(GO:0034629) |
| 0.0 |
0.6 |
GO:0045747 |
positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 |
0.1 |
GO:0003376 |
sphingosine-1-phosphate signaling pathway(GO:0003376) sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 |
0.1 |
GO:1900060 |
negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.0 |
0.2 |
GO:0060081 |
membrane hyperpolarization(GO:0060081) |
| 0.0 |
0.5 |
GO:0032008 |
positive regulation of TOR signaling(GO:0032008) |
| 0.0 |
0.2 |
GO:0030042 |
actin filament depolymerization(GO:0030042) |
| 0.0 |
0.7 |
GO:0035329 |
hippo signaling(GO:0035329) |
| 0.0 |
0.1 |
GO:0070458 |
detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 0.0 |
0.1 |
GO:0030327 |
prenylated protein catabolic process(GO:0030327) |
| 0.0 |
0.1 |
GO:0038018 |
Wnt receptor catabolic process(GO:0038018) |
| 0.0 |
0.1 |
GO:0002036 |
regulation of L-glutamate transport(GO:0002036) negative regulation of mitochondrial membrane potential(GO:0010917) |
| 0.0 |
0.1 |
GO:2000809 |
positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.0 |
0.2 |
GO:0010801 |
negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.0 |
0.2 |
GO:0042059 |
negative regulation of epidermal growth factor receptor signaling pathway(GO:0042059) |
| 0.0 |
0.1 |
GO:0006867 |
asparagine transport(GO:0006867) glutamine transport(GO:0006868) |
| 0.0 |
0.2 |
GO:0060736 |
prostate gland growth(GO:0060736) |
| 0.0 |
0.2 |
GO:0018126 |
protein hydroxylation(GO:0018126) |
| 0.0 |
0.4 |
GO:0001706 |
endoderm formation(GO:0001706) |
| 0.0 |
0.2 |
GO:0002115 |
store-operated calcium entry(GO:0002115) |
| 0.0 |
0.1 |
GO:0018026 |
peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 |
0.3 |
GO:2000001 |
regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 |
0.2 |
GO:0032780 |
negative regulation of ATPase activity(GO:0032780) |
| 0.0 |
0.1 |
GO:0030578 |
PML body organization(GO:0030578) |
| 0.0 |
0.2 |
GO:0008535 |
respiratory chain complex IV assembly(GO:0008535) |
| 0.0 |
0.4 |
GO:0045022 |
early endosome to late endosome transport(GO:0045022) |
| 0.0 |
0.4 |
GO:0048791 |
calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 |
0.8 |
GO:0048663 |
neuron fate commitment(GO:0048663) |
| 0.0 |
0.7 |
GO:0001541 |
ovarian follicle development(GO:0001541) |
| 0.0 |
0.1 |
GO:0014842 |
regulation of skeletal muscle satellite cell proliferation(GO:0014842) |
| 0.0 |
0.2 |
GO:0000132 |
establishment of mitotic spindle orientation(GO:0000132) |
| 0.0 |
0.3 |
GO:0032288 |
myelin assembly(GO:0032288) |
| 0.0 |
0.1 |
GO:0098903 |
regulation of membrane repolarization during action potential(GO:0098903) |
| 0.0 |
0.6 |
GO:0007405 |
neuroblast proliferation(GO:0007405) |
| 0.0 |
0.5 |
GO:0048013 |
ephrin receptor signaling pathway(GO:0048013) |
| 0.0 |
0.1 |
GO:0020027 |
hemoglobin metabolic process(GO:0020027) |
| 0.0 |
0.5 |
GO:0043388 |
positive regulation of DNA binding(GO:0043388) |
| 0.0 |
0.1 |
GO:0035735 |
intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 |
0.2 |
GO:0032897 |
negative regulation of viral transcription(GO:0032897) |
| 0.0 |
0.5 |
GO:0070373 |
negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.0 |
0.1 |
GO:1901409 |
regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 |
0.4 |
GO:0046579 |
positive regulation of Ras protein signal transduction(GO:0046579) |
| 0.0 |
0.0 |
GO:0046077 |
dUDP biosynthetic process(GO:0006227) pyrimidine nucleoside diphosphate biosynthetic process(GO:0009139) pyrimidine deoxyribonucleoside diphosphate metabolic process(GO:0009196) pyrimidine deoxyribonucleoside diphosphate biosynthetic process(GO:0009197) dUDP metabolic process(GO:0046077) |
| 0.0 |
0.1 |
GO:0033599 |
regulation of mammary gland epithelial cell proliferation(GO:0033599) |
| 0.0 |
0.3 |
GO:0016578 |
histone deubiquitination(GO:0016578) |