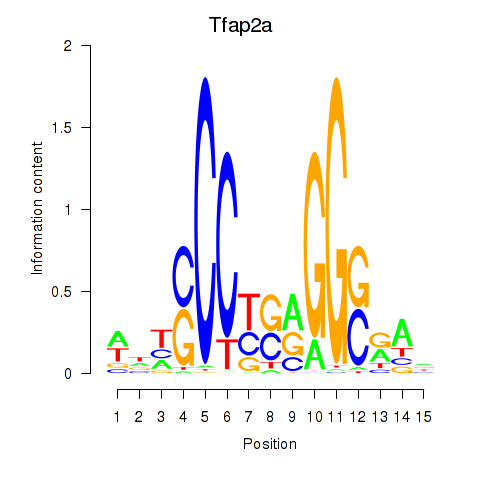
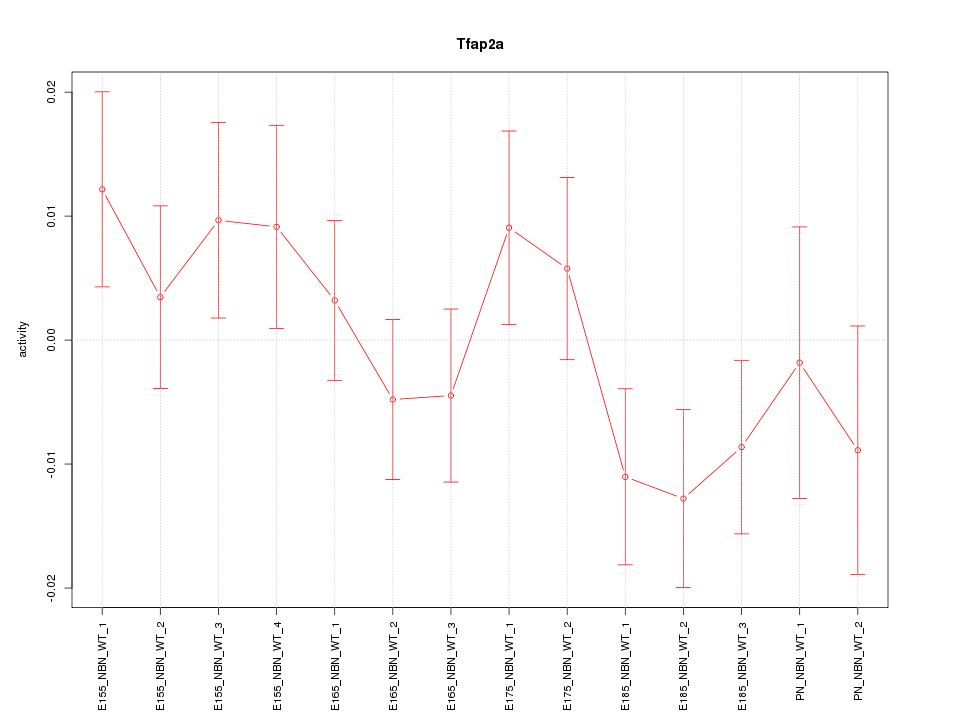
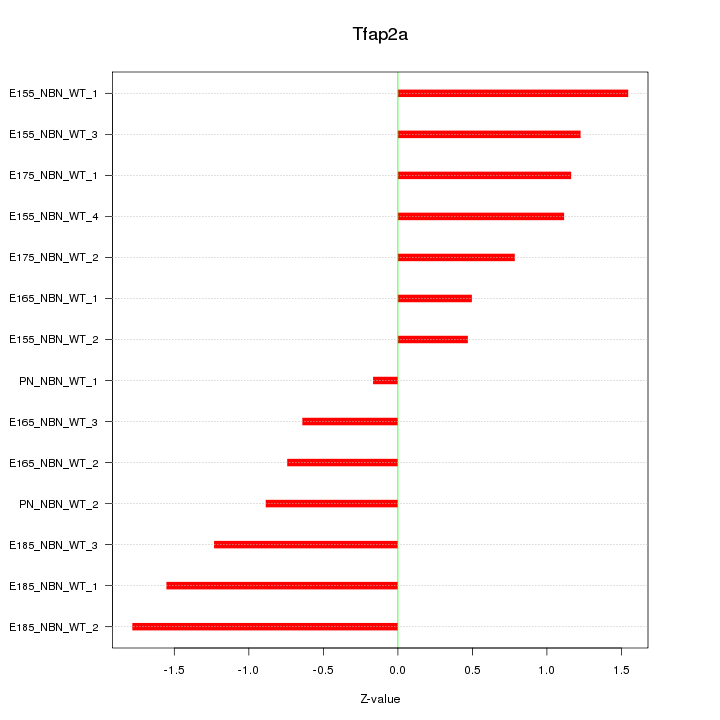
| 0.7 |
2.9 |
GO:0033024 |
mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) |
| 0.6 |
1.7 |
GO:0060598 |
dichotomous subdivision of terminal units involved in mammary gland duct morphogenesis(GO:0060598) |
| 0.5 |
1.6 |
GO:0045976 |
negative regulation of mitotic cell cycle, embryonic(GO:0045976) |
| 0.3 |
0.9 |
GO:0042271 |
susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.3 |
1.2 |
GO:0014846 |
esophagus smooth muscle contraction(GO:0014846) |
| 0.3 |
0.9 |
GO:0070973 |
protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.3 |
2.2 |
GO:0035542 |
regulation of SNARE complex assembly(GO:0035542) |
| 0.3 |
0.8 |
GO:0048686 |
regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 0.3 |
0.8 |
GO:0008228 |
opsonization(GO:0008228) |
| 0.3 |
5.1 |
GO:0050650 |
chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.3 |
1.3 |
GO:0002121 |
inter-male aggressive behavior(GO:0002121) |
| 0.2 |
0.7 |
GO:0061535 |
glutamate secretion, neurotransmission(GO:0061535) |
| 0.2 |
0.6 |
GO:0048211 |
Golgi vesicle docking(GO:0048211) |
| 0.2 |
0.6 |
GO:0045110 |
intermediate filament bundle assembly(GO:0045110) |
| 0.2 |
0.4 |
GO:1904192 |
cholangiocyte apoptotic process(GO:1902488) regulation of cholangiocyte apoptotic process(GO:1904192) negative regulation of cholangiocyte apoptotic process(GO:1904193) |
| 0.2 |
1.1 |
GO:0060017 |
parathyroid gland development(GO:0060017) |
| 0.2 |
1.7 |
GO:0030322 |
stabilization of membrane potential(GO:0030322) |
| 0.2 |
0.2 |
GO:0061324 |
canonical Wnt signaling pathway involved in positive regulation of cardiac outflow tract cell proliferation(GO:0061324) regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901963) |
| 0.2 |
0.5 |
GO:0046959 |
habituation(GO:0046959) |
| 0.2 |
0.7 |
GO:0035795 |
negative regulation of mitochondrial membrane permeability(GO:0035795) |
| 0.2 |
0.5 |
GO:0060060 |
post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.1 |
0.7 |
GO:0061086 |
negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.1 |
1.8 |
GO:0060052 |
neurofilament cytoskeleton organization(GO:0060052) |
| 0.1 |
0.6 |
GO:0033326 |
cerebrospinal fluid secretion(GO:0033326) |
| 0.1 |
0.6 |
GO:0019355 |
nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.1 |
0.7 |
GO:0016266 |
O-glycan processing(GO:0016266) |
| 0.1 |
0.4 |
GO:0021577 |
hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.1 |
0.5 |
GO:0048304 |
positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.1 |
0.5 |
GO:1904694 |
negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.1 |
0.5 |
GO:1901204 |
positive regulation of guanylate cyclase activity(GO:0031284) regulation of adrenergic receptor signaling pathway involved in heart process(GO:1901204) |
| 0.1 |
0.5 |
GO:0032289 |
central nervous system myelin formation(GO:0032289) cardiac cell fate specification(GO:0060912) |
| 0.1 |
0.5 |
GO:0061198 |
fungiform papilla formation(GO:0061198) |
| 0.1 |
0.6 |
GO:0007223 |
Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.1 |
0.6 |
GO:0051012 |
microtubule sliding(GO:0051012) negative regulation of nonmotile primary cilium assembly(GO:1902856) |
| 0.1 |
0.8 |
GO:0048680 |
positive regulation of axon regeneration(GO:0048680) |
| 0.1 |
0.7 |
GO:1902897 |
regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.1 |
0.6 |
GO:1903361 |
protein localization to basolateral plasma membrane(GO:1903361) |
| 0.1 |
0.6 |
GO:0044314 |
protein K27-linked ubiquitination(GO:0044314) |
| 0.1 |
1.5 |
GO:0070842 |
aggresome assembly(GO:0070842) |
| 0.1 |
0.3 |
GO:0015712 |
hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) phosphate ion transmembrane transport(GO:0035435) |
| 0.1 |
0.4 |
GO:0006203 |
dGTP catabolic process(GO:0006203) dATP catabolic process(GO:0046061) |
| 0.1 |
0.3 |
GO:0061588 |
calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylserine scrambling(GO:0061589) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.1 |
0.4 |
GO:0051088 |
PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.1 |
1.2 |
GO:0033572 |
transferrin transport(GO:0033572) |
| 0.1 |
0.3 |
GO:1904431 |
positive regulation of t-circle formation(GO:1904431) |
| 0.1 |
0.3 |
GO:0019401 |
alditol biosynthetic process(GO:0019401) |
| 0.1 |
0.8 |
GO:2000857 |
positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.1 |
1.6 |
GO:0050832 |
defense response to fungus(GO:0050832) |
| 0.1 |
0.4 |
GO:0045343 |
MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) positive regulation of MHC class I biosynthetic process(GO:0045345) |
| 0.1 |
0.3 |
GO:0060282 |
positive regulation of oocyte development(GO:0060282) |
| 0.1 |
0.3 |
GO:0036092 |
phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 |
0.4 |
GO:2000211 |
regulation of glutamate metabolic process(GO:2000211) |
| 0.1 |
0.3 |
GO:0006109 |
regulation of carbohydrate metabolic process(GO:0006109) |
| 0.1 |
0.4 |
GO:0007256 |
activation of JNKK activity(GO:0007256) |
| 0.1 |
0.3 |
GO:0007056 |
spindle assembly involved in female meiosis(GO:0007056) |
| 0.1 |
0.5 |
GO:0060046 |
regulation of acrosome reaction(GO:0060046) |
| 0.1 |
0.3 |
GO:0035621 |
ER to Golgi ceramide transport(GO:0035621) ceramide transport(GO:0035627) |
| 0.1 |
0.4 |
GO:0015867 |
ATP transport(GO:0015867) |
| 0.1 |
0.4 |
GO:0000414 |
regulation of histone H3-K36 methylation(GO:0000414) |
| 0.1 |
0.2 |
GO:0070602 |
regulation of centromeric sister chromatid cohesion(GO:0070602) |
| 0.1 |
0.5 |
GO:1990118 |
sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 |
0.3 |
GO:0002175 |
protein localization to paranode region of axon(GO:0002175) |
| 0.1 |
2.3 |
GO:0006198 |
cAMP catabolic process(GO:0006198) |
| 0.1 |
0.3 |
GO:0090234 |
regulation of kinetochore assembly(GO:0090234) |
| 0.1 |
0.7 |
GO:1900112 |
regulation of histone H3-K9 trimethylation(GO:1900112) negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.1 |
0.3 |
GO:0006369 |
termination of RNA polymerase II transcription(GO:0006369) |
| 0.1 |
0.3 |
GO:0046501 |
protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.1 |
1.5 |
GO:0051654 |
establishment of mitochondrion localization(GO:0051654) |
| 0.1 |
0.3 |
GO:0015692 |
vanadium ion transport(GO:0015676) lead ion transport(GO:0015692) |
| 0.1 |
0.2 |
GO:0045041 |
protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.1 |
0.3 |
GO:2001032 |
regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.1 |
0.6 |
GO:0018230 |
peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 |
0.3 |
GO:0055071 |
cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.1 |
0.9 |
GO:0035020 |
regulation of Rac protein signal transduction(GO:0035020) |
| 0.1 |
0.3 |
GO:0060178 |
regulation of exocyst localization(GO:0060178) |
| 0.1 |
0.2 |
GO:0072675 |
osteoclast fusion(GO:0072675) |
| 0.1 |
0.3 |
GO:1900122 |
positive regulation of receptor binding(GO:1900122) |
| 0.1 |
0.1 |
GO:0021933 |
radial glia guided migration of cerebellar granule cell(GO:0021933) |
| 0.1 |
1.0 |
GO:0070884 |
regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.1 |
0.3 |
GO:0033762 |
response to glucagon(GO:0033762) |
| 0.1 |
0.3 |
GO:0070125 |
mitochondrial translational elongation(GO:0070125) |
| 0.1 |
0.1 |
GO:0030997 |
regulation of centriole-centriole cohesion(GO:0030997) |
| 0.1 |
0.2 |
GO:1902528 |
regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.1 |
0.4 |
GO:1903551 |
regulation of extracellular exosome assembly(GO:1903551) |
| 0.1 |
0.5 |
GO:1903608 |
protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.1 |
0.2 |
GO:0051902 |
negative regulation of mitochondrial depolarization(GO:0051902) negative regulation of membrane depolarization(GO:1904180) |
| 0.1 |
0.3 |
GO:0009235 |
cobalamin metabolic process(GO:0009235) |
| 0.1 |
0.2 |
GO:1903054 |
lysosomal membrane organization(GO:0097212) negative regulation of extracellular matrix organization(GO:1903054) positive regulation of protein folding(GO:1903334) |
| 0.1 |
0.2 |
GO:0097500 |
receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.1 |
0.2 |
GO:0030300 |
regulation of intestinal cholesterol absorption(GO:0030300) |
| 0.1 |
1.9 |
GO:0036075 |
endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.1 |
0.3 |
GO:1905049 |
negative regulation of metalloendopeptidase activity(GO:1904684) negative regulation of metallopeptidase activity(GO:1905049) |
| 0.0 |
0.4 |
GO:0061368 |
behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.0 |
0.2 |
GO:2000483 |
negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.0 |
0.7 |
GO:0060213 |
regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.0 |
0.2 |
GO:0046726 |
positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.0 |
0.5 |
GO:0033148 |
positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.0 |
0.1 |
GO:0009446 |
putrescine biosynthetic process(GO:0009446) |
| 0.0 |
0.3 |
GO:0016584 |
nucleosome positioning(GO:0016584) |
| 0.0 |
0.2 |
GO:0071442 |
positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.0 |
0.1 |
GO:1904177 |
regulation of adipose tissue development(GO:1904177) |
| 0.0 |
0.1 |
GO:0030397 |
membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.0 |
0.2 |
GO:0071034 |
CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.0 |
0.9 |
GO:0006047 |
UDP-N-acetylglucosamine metabolic process(GO:0006047) |
| 0.0 |
0.2 |
GO:0071955 |
recycling endosome to Golgi transport(GO:0071955) |
| 0.0 |
0.1 |
GO:0036091 |
positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.0 |
0.6 |
GO:0048172 |
regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 |
0.2 |
GO:0021698 |
cerebellar cortex structural organization(GO:0021698) |
| 0.0 |
0.1 |
GO:0019858 |
cytosine metabolic process(GO:0019858) |
| 0.0 |
0.6 |
GO:0097284 |
hepatocyte apoptotic process(GO:0097284) |
| 0.0 |
0.3 |
GO:0045078 |
enucleate erythrocyte differentiation(GO:0043353) positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.0 |
0.8 |
GO:0046854 |
phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 |
0.1 |
GO:0070829 |
heterochromatin maintenance(GO:0070829) |
| 0.0 |
0.2 |
GO:0007197 |
adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.0 |
0.1 |
GO:0060368 |
regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060368) |
| 0.0 |
0.4 |
GO:0033008 |
positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.0 |
0.5 |
GO:0007340 |
acrosome reaction(GO:0007340) |
| 0.0 |
0.8 |
GO:2000651 |
positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 |
0.1 |
GO:0010985 |
negative regulation of lipoprotein particle clearance(GO:0010985) |
| 0.0 |
0.3 |
GO:0051574 |
positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 |
0.1 |
GO:0034454 |
microtubule anchoring at centrosome(GO:0034454) |
| 0.0 |
0.5 |
GO:0071447 |
cellular response to hydroperoxide(GO:0071447) |
| 0.0 |
0.8 |
GO:0008340 |
determination of adult lifespan(GO:0008340) |
| 0.0 |
0.5 |
GO:0008209 |
androgen metabolic process(GO:0008209) |
| 0.0 |
0.6 |
GO:0003338 |
metanephros morphogenesis(GO:0003338) |
| 0.0 |
0.2 |
GO:0071557 |
histone H3-K27 demethylation(GO:0071557) |
| 0.0 |
0.8 |
GO:0046856 |
phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 |
0.2 |
GO:0017196 |
N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 |
0.2 |
GO:0033564 |
anterior/posterior axon guidance(GO:0033564) |
| 0.0 |
0.3 |
GO:0051639 |
actin filament network formation(GO:0051639) |
| 0.0 |
0.1 |
GO:1904354 |
negative regulation of telomere capping(GO:1904354) |
| 0.0 |
0.1 |
GO:0014719 |
skeletal muscle satellite cell activation(GO:0014719) |
| 0.0 |
0.1 |
GO:0060011 |
Sertoli cell proliferation(GO:0060011) |
| 0.0 |
0.1 |
GO:0045654 |
positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.0 |
0.2 |
GO:0021775 |
smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.0 |
0.3 |
GO:0032876 |
negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 |
0.4 |
GO:0003215 |
cardiac right ventricle morphogenesis(GO:0003215) |
| 0.0 |
0.7 |
GO:0007020 |
microtubule nucleation(GO:0007020) |
| 0.0 |
0.2 |
GO:0071318 |
cellular response to ATP(GO:0071318) |
| 0.0 |
0.1 |
GO:1902035 |
positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.0 |
0.1 |
GO:1990253 |
cellular response to leucine starvation(GO:1990253) |
| 0.0 |
0.2 |
GO:0090209 |
negative regulation of triglyceride metabolic process(GO:0090209) |
| 0.0 |
0.3 |
GO:0033173 |
calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 |
0.3 |
GO:0034773 |
histone H4-K20 trimethylation(GO:0034773) |
| 0.0 |
0.5 |
GO:0035873 |
lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.0 |
0.9 |
GO:2000171 |
negative regulation of dendrite development(GO:2000171) |
| 0.0 |
0.1 |
GO:0006680 |
glucosylceramide catabolic process(GO:0006680) |
| 0.0 |
0.1 |
GO:0043553 |
negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 |
0.2 |
GO:0045974 |
miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 |
0.3 |
GO:0097034 |
mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 |
0.2 |
GO:1903300 |
negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 |
0.0 |
GO:0070827 |
chromatin maintenance(GO:0070827) |
| 0.0 |
0.1 |
GO:0019676 |
ammonia assimilation cycle(GO:0019676) |
| 0.0 |
0.0 |
GO:0061030 |
epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.0 |
0.1 |
GO:0016259 |
selenocysteine metabolic process(GO:0016259) |
| 0.0 |
0.5 |
GO:0021702 |
cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 |
0.0 |
GO:1903377 |
negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 |
0.1 |
GO:0089711 |
L-glutamate transmembrane transport(GO:0089711) |
| 0.0 |
0.4 |
GO:0018345 |
protein palmitoylation(GO:0018345) |
| 0.0 |
0.2 |
GO:0043433 |
negative regulation of sequence-specific DNA binding transcription factor activity(GO:0043433) |
| 0.0 |
0.1 |
GO:0006488 |
dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 |
0.5 |
GO:0044030 |
regulation of DNA methylation(GO:0044030) |
| 0.0 |
0.1 |
GO:0030862 |
neuroblast division in subventricular zone(GO:0021849) regulation of polarized epithelial cell differentiation(GO:0030860) positive regulation of polarized epithelial cell differentiation(GO:0030862) |
| 0.0 |
0.2 |
GO:0033299 |
secretion of lysosomal enzymes(GO:0033299) |
| 0.0 |
0.4 |
GO:0099643 |
neurotransmitter secretion(GO:0007269) presynaptic process involved in chemical synaptic transmission(GO:0099531) signal release from synapse(GO:0099643) |
| 0.0 |
0.5 |
GO:0003301 |
physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 |
0.2 |
GO:0031268 |
pseudopodium organization(GO:0031268) |
| 0.0 |
0.1 |
GO:0002901 |
mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.0 |
0.5 |
GO:0061098 |
positive regulation of protein tyrosine kinase activity(GO:0061098) |
| 0.0 |
0.3 |
GO:0035036 |
binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) |
| 0.0 |
0.1 |
GO:0045163 |
clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 |
0.2 |
GO:0007250 |
activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 |
0.1 |
GO:0006904 |
vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 |
0.5 |
GO:0045620 |
negative regulation of lymphocyte differentiation(GO:0045620) |
| 0.0 |
0.1 |
GO:1904996 |
diapedesis(GO:0050904) positive regulation of leukocyte adhesion to vascular endothelial cell(GO:1904996) |
| 0.0 |
0.2 |
GO:1990126 |
retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 |
0.8 |
GO:0060395 |
SMAD protein signal transduction(GO:0060395) |
| 0.0 |
0.0 |
GO:0048162 |
preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.0 |
0.1 |
GO:0042135 |
neurotransmitter catabolic process(GO:0042135) |
| 0.0 |
0.1 |
GO:0043922 |
negative regulation by host of viral transcription(GO:0043922) |
| 0.0 |
0.2 |
GO:0040037 |
negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.0 |
0.2 |
GO:0010715 |
regulation of extracellular matrix disassembly(GO:0010715) |
| 0.0 |
0.2 |
GO:1900037 |
regulation of cellular response to hypoxia(GO:1900037) |
| 0.0 |
0.3 |
GO:0048147 |
negative regulation of fibroblast proliferation(GO:0048147) |
| 0.0 |
0.7 |
GO:0019319 |
hexose biosynthetic process(GO:0019319) |
| 0.0 |
0.3 |
GO:1902236 |
negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.0 |
0.4 |
GO:0051966 |
regulation of synaptic transmission, glutamatergic(GO:0051966) |
| 0.0 |
0.1 |
GO:0090220 |
meiotic telomere clustering(GO:0045141) chromosome localization to nuclear envelope involved in homologous chromosome segregation(GO:0090220) |
| 0.0 |
0.2 |
GO:0071385 |
cellular response to glucocorticoid stimulus(GO:0071385) |
| 0.0 |
0.6 |
GO:0051897 |
positive regulation of protein kinase B signaling(GO:0051897) |
| 0.0 |
0.1 |
GO:0003334 |
keratinocyte development(GO:0003334) |
| 0.0 |
0.1 |
GO:0045187 |
regulation of circadian sleep/wake cycle(GO:0042749) regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.0 |
0.1 |
GO:0035269 |
protein O-linked mannosylation(GO:0035269) |
| 0.0 |
0.2 |
GO:0045793 |
positive regulation of cell size(GO:0045793) |
| 0.0 |
0.2 |
GO:0010971 |
positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 0.0 |
0.7 |
GO:0000245 |
spliceosomal complex assembly(GO:0000245) |
| 0.0 |
0.0 |
GO:1903726 |
negative regulation of phospholipid biosynthetic process(GO:0071072) negative regulation of phospholipid metabolic process(GO:1903726) regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.0 |
0.1 |
GO:0002021 |
response to dietary excess(GO:0002021) |
| 0.0 |
0.8 |
GO:0002028 |
regulation of sodium ion transport(GO:0002028) |
| 0.0 |
0.1 |
GO:0090161 |
Golgi ribbon formation(GO:0090161) |
| 0.0 |
0.4 |
GO:0006378 |
mRNA polyadenylation(GO:0006378) |
| 0.0 |
1.4 |
GO:0071804 |
cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 |
0.3 |
GO:0007205 |
protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 |
0.2 |
GO:0031100 |
organ regeneration(GO:0031100) |
| 0.0 |
0.1 |
GO:0010825 |
positive regulation of centrosome duplication(GO:0010825) |
| 0.0 |
0.0 |
GO:0061092 |
regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.0 |
0.2 |
GO:2000310 |
regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.0 |
0.3 |
GO:0043001 |
Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 |
0.1 |
GO:0002317 |
plasma cell differentiation(GO:0002317) |
| 0.0 |
0.1 |
GO:0010603 |
regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |