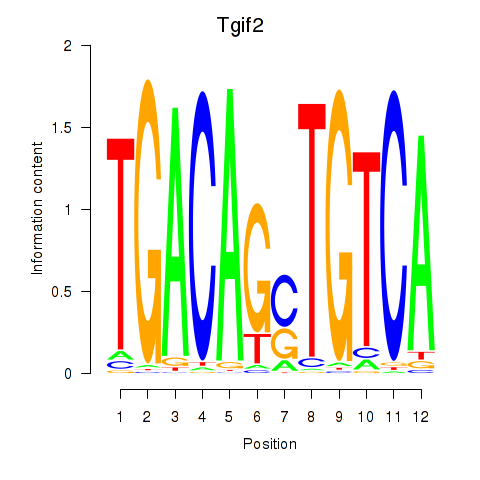
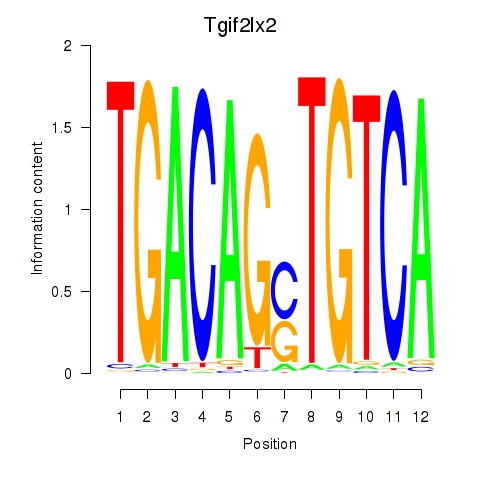
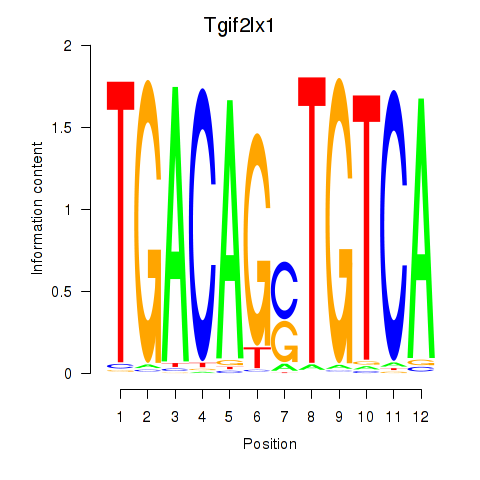
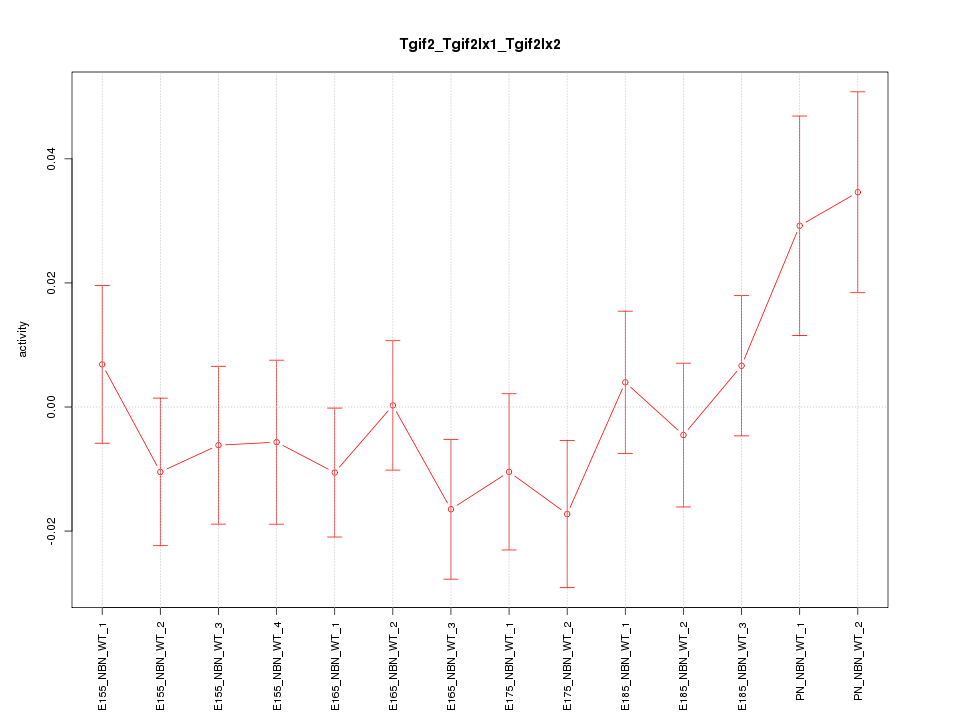
| 1.3 |
5.2 |
GO:1903181 |
regulation of dopamine biosynthetic process(GO:1903179) positive regulation of dopamine biosynthetic process(GO:1903181) |
| 1.2 |
5.8 |
GO:0021905 |
pancreatic A cell development(GO:0003322) forebrain-midbrain boundary formation(GO:0021905) somatic motor neuron fate commitment(GO:0021917) regulation of transcription from RNA polymerase II promoter involved in somatic motor neuron fate commitment(GO:0021918) |
| 0.4 |
1.6 |
GO:0006842 |
tricarboxylic acid transport(GO:0006842) succinate transport(GO:0015744) citrate transport(GO:0015746) |
| 0.3 |
1.0 |
GO:0006030 |
chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.2 |
1.6 |
GO:0070092 |
regulation of glucagon secretion(GO:0070092) |
| 0.2 |
1.0 |
GO:0034421 |
post-translational protein acetylation(GO:0034421) |
| 0.1 |
1.2 |
GO:0046459 |
short-chain fatty acid metabolic process(GO:0046459) |
| 0.1 |
1.4 |
GO:0035999 |
tetrahydrofolate interconversion(GO:0035999) |
| 0.1 |
1.5 |
GO:0007096 |
regulation of exit from mitosis(GO:0007096) |
| 0.1 |
0.6 |
GO:2000042 |
negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.1 |
2.3 |
GO:0001504 |
neurotransmitter uptake(GO:0001504) |
| 0.1 |
0.6 |
GO:0032911 |
nerve growth factor production(GO:0032902) negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.1 |
0.3 |
GO:0060023 |
soft palate development(GO:0060023) |
| 0.1 |
1.9 |
GO:1904816 |
positive regulation of protein localization to chromosome, telomeric region(GO:1904816) |
| 0.1 |
1.7 |
GO:0050901 |
leukocyte tethering or rolling(GO:0050901) |
| 0.1 |
0.4 |
GO:0042796 |
snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.1 |
0.5 |
GO:0042701 |
progesterone secretion(GO:0042701) |
| 0.1 |
0.7 |
GO:0048478 |
replication fork protection(GO:0048478) |
| 0.1 |
0.3 |
GO:0045204 |
MAPK export from nucleus(GO:0045204) |
| 0.1 |
1.0 |
GO:0060081 |
membrane hyperpolarization(GO:0060081) |
| 0.1 |
0.5 |
GO:0006177 |
GMP biosynthetic process(GO:0006177) |
| 0.1 |
0.5 |
GO:0002591 |
positive regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002591) |
| 0.1 |
0.3 |
GO:1902268 |
negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.1 |
0.2 |
GO:0021852 |
pyramidal neuron migration(GO:0021852) |
| 0.1 |
0.4 |
GO:0034316 |
negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.1 |
0.5 |
GO:0035845 |
photoreceptor cell outer segment organization(GO:0035845) |
| 0.1 |
1.6 |
GO:0070584 |
mitochondrion morphogenesis(GO:0070584) |
| 0.1 |
0.6 |
GO:0070886 |
positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.1 |
0.2 |
GO:2001187 |
positive regulation of CD8-positive, alpha-beta T cell activation(GO:2001187) |
| 0.1 |
1.3 |
GO:0071108 |
protein K48-linked deubiquitination(GO:0071108) |
| 0.1 |
1.5 |
GO:0002474 |
antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 |
0.3 |
GO:0002317 |
plasma cell differentiation(GO:0002317) |
| 0.0 |
0.6 |
GO:0019886 |
antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.0 |
0.4 |
GO:0046415 |
urate metabolic process(GO:0046415) |
| 0.0 |
0.5 |
GO:0051103 |
DNA ligation involved in DNA repair(GO:0051103) |
| 0.0 |
1.1 |
GO:0030224 |
monocyte differentiation(GO:0030224) |
| 0.0 |
1.1 |
GO:0006730 |
one-carbon metabolic process(GO:0006730) |
| 0.0 |
0.3 |
GO:0006451 |
selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 |
1.5 |
GO:0007157 |
heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 |
0.8 |
GO:0007064 |
mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 |
0.3 |
GO:0019800 |
peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 |
1.7 |
GO:0030901 |
midbrain development(GO:0030901) |
| 0.0 |
1.0 |
GO:0031365 |
N-terminal protein amino acid modification(GO:0031365) |
| 0.0 |
0.6 |
GO:0040034 |
regulation of development, heterochronic(GO:0040034) |
| 0.0 |
0.1 |
GO:0048239 |
negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 0.0 |
0.3 |
GO:0097284 |
hepatocyte apoptotic process(GO:0097284) |
| 0.0 |
0.7 |
GO:0008608 |
attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.0 |
0.1 |
GO:0015808 |
L-alanine transport(GO:0015808) |
| 0.0 |
0.2 |
GO:1901621 |
negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 |
0.1 |
GO:0010908 |
regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 |
1.5 |
GO:0042698 |
ovulation cycle(GO:0042698) |
| 0.0 |
0.2 |
GO:0036376 |
sodium ion export from cell(GO:0036376) |
| 0.0 |
0.3 |
GO:2001014 |
regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.0 |
0.9 |
GO:0035690 |
cellular response to drug(GO:0035690) |
| 0.0 |
0.2 |
GO:0045187 |
regulation of circadian sleep/wake cycle(GO:0042749) regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.0 |
0.1 |
GO:0009146 |
purine nucleoside triphosphate catabolic process(GO:0009146) |
| 0.0 |
0.3 |
GO:0045956 |
positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.0 |
0.2 |
GO:0010867 |
positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.0 |
0.2 |
GO:0042407 |
cristae formation(GO:0042407) |
| 0.0 |
0.1 |
GO:0018230 |
peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 |
0.4 |
GO:0031648 |
protein destabilization(GO:0031648) |
| 0.0 |
0.6 |
GO:0042475 |
odontogenesis of dentin-containing tooth(GO:0042475) |