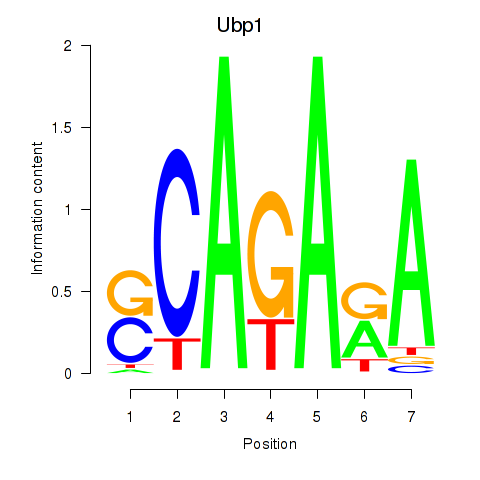
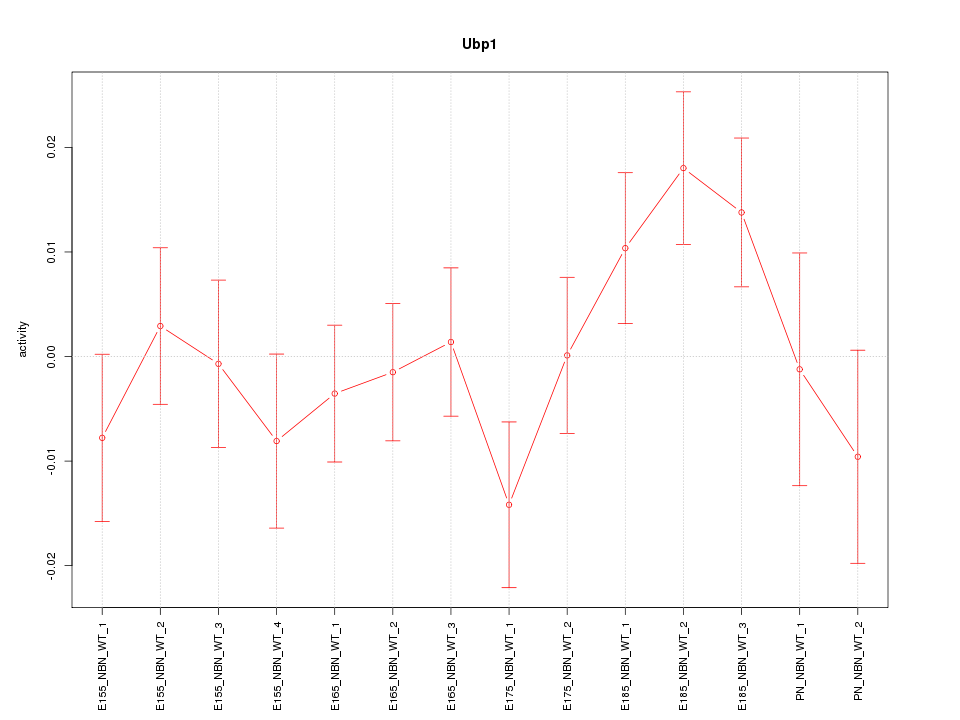
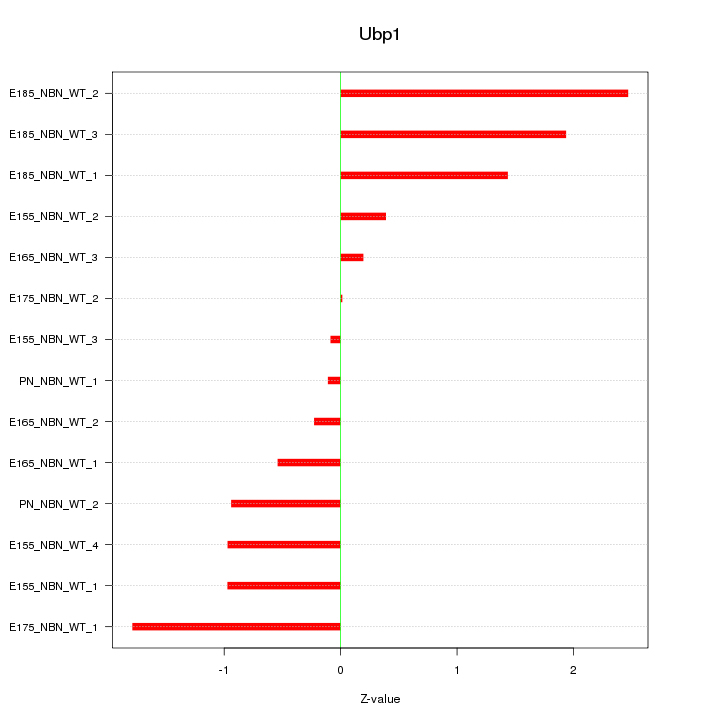
| 0.7 |
2.2 |
GO:0042196 |
dichloromethane metabolic process(GO:0018900) chlorinated hydrocarbon metabolic process(GO:0042196) halogenated hydrocarbon metabolic process(GO:0042197) |
| 0.4 |
2.2 |
GO:0002457 |
T cell antigen processing and presentation(GO:0002457) |
| 0.4 |
2.0 |
GO:0045347 |
negative regulation of MHC class II biosynthetic process(GO:0045347) negative regulation of cytolysis(GO:0045918) |
| 0.2 |
0.7 |
GO:0008228 |
opsonization(GO:0008228) modulation of molecular function in other organism(GO:0044359) negative regulation of molecular function in other organism(GO:0044362) negative regulation of molecular function in other organism involved in symbiotic interaction(GO:0052204) modulation of molecular function in other organism involved in symbiotic interaction(GO:0052205) negative regulation by host of symbiont molecular function(GO:0052405) modification by host of symbiont molecular function(GO:0052428) |
| 0.2 |
0.5 |
GO:0060166 |
olfactory pit development(GO:0060166) |
| 0.2 |
0.5 |
GO:0030421 |
defecation(GO:0030421) |
| 0.2 |
0.5 |
GO:0010986 |
positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.2 |
0.5 |
GO:0035790 |
platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) |
| 0.2 |
1.1 |
GO:0019370 |
leukotriene biosynthetic process(GO:0019370) |
| 0.1 |
0.6 |
GO:0048539 |
bone marrow development(GO:0048539) |
| 0.1 |
0.4 |
GO:0071873 |
response to norepinephrine(GO:0071873) |
| 0.1 |
0.4 |
GO:0003100 |
regulation of systemic arterial blood pressure by endothelin(GO:0003100) vein smooth muscle contraction(GO:0014826) |
| 0.1 |
0.7 |
GO:0046598 |
positive regulation of viral entry into host cell(GO:0046598) |
| 0.1 |
0.7 |
GO:1903026 |
negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.1 |
0.4 |
GO:0003032 |
detection of oxygen(GO:0003032) regulation of epithelial to mesenchymal transition involved in endocardial cushion formation(GO:1905005) |
| 0.1 |
0.9 |
GO:0097460 |
ferrous iron import into cell(GO:0097460) |
| 0.1 |
0.9 |
GO:0071361 |
cellular response to ethanol(GO:0071361) |
| 0.1 |
0.4 |
GO:0060217 |
hemangioblast cell differentiation(GO:0060217) |
| 0.1 |
0.4 |
GO:0001193 |
maintenance of transcriptional fidelity during DNA-templated transcription elongation(GO:0001192) maintenance of transcriptional fidelity during DNA-templated transcription elongation from RNA polymerase II promoter(GO:0001193) |
| 0.1 |
0.4 |
GO:2000569 |
T-helper 2 cell activation(GO:0035712) positive regulation of T-helper 17 type immune response(GO:2000318) regulation of T-helper 2 cell activation(GO:2000569) positive regulation of T-helper 2 cell activation(GO:2000570) |
| 0.1 |
0.4 |
GO:0051695 |
actin filament uncapping(GO:0051695) |
| 0.1 |
0.7 |
GO:0033227 |
dsRNA transport(GO:0033227) |
| 0.1 |
0.3 |
GO:0046078 |
deoxyribonucleoside monophosphate biosynthetic process(GO:0009157) pyrimidine deoxyribonucleoside monophosphate biosynthetic process(GO:0009177) pyrimidine deoxyribonucleotide catabolic process(GO:0009223) dUMP metabolic process(GO:0046078) |
| 0.1 |
0.3 |
GO:1904569 |
regulation of selenocysteine incorporation(GO:1904569) |
| 0.1 |
1.0 |
GO:0019800 |
peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 |
0.3 |
GO:0071374 |
cellular response to parathyroid hormone stimulus(GO:0071374) |
| 0.1 |
0.6 |
GO:0006123 |
mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 |
0.3 |
GO:0001869 |
regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 0.1 |
0.3 |
GO:0035519 |
protein K29-linked ubiquitination(GO:0035519) |
| 0.1 |
0.4 |
GO:0007412 |
axon target recognition(GO:0007412) |
| 0.1 |
0.4 |
GO:0071726 |
response to diacyl bacterial lipopeptide(GO:0071724) cellular response to diacyl bacterial lipopeptide(GO:0071726) |
| 0.1 |
0.3 |
GO:0007181 |
transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.1 |
0.2 |
GO:0035990 |
tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.1 |
0.8 |
GO:0060770 |
negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.1 |
0.3 |
GO:0019478 |
D-amino acid catabolic process(GO:0019478) |
| 0.1 |
0.5 |
GO:0030644 |
cellular chloride ion homeostasis(GO:0030644) |
| 0.1 |
1.5 |
GO:0042182 |
ketone catabolic process(GO:0042182) |
| 0.1 |
0.3 |
GO:0051311 |
spindle assembly involved in female meiosis(GO:0007056) meiotic metaphase plate congression(GO:0051311) |
| 0.1 |
0.5 |
GO:0021797 |
forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.1 |
0.4 |
GO:0030091 |
protein repair(GO:0030091) |
| 0.1 |
0.6 |
GO:0055098 |
response to low-density lipoprotein particle(GO:0055098) |
| 0.1 |
1.0 |
GO:0097067 |
cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.1 |
0.2 |
GO:0019405 |
hexitol metabolic process(GO:0006059) alditol catabolic process(GO:0019405) fructose biosynthetic process(GO:0046370) |
| 0.1 |
0.1 |
GO:0002322 |
B cell proliferation involved in immune response(GO:0002322) |
| 0.1 |
0.1 |
GO:0034310 |
primary alcohol catabolic process(GO:0034310) |
| 0.1 |
0.3 |
GO:0097494 |
positive regulation of neurotransmitter uptake(GO:0051582) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) regulation of vesicle size(GO:0097494) |
| 0.1 |
0.7 |
GO:2001214 |
positive regulation of vasculogenesis(GO:2001214) |
| 0.1 |
0.7 |
GO:0009404 |
toxin metabolic process(GO:0009404) |
| 0.1 |
0.7 |
GO:0034551 |
respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.1 |
0.4 |
GO:0036444 |
calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.1 |
0.2 |
GO:2000987 |
positive regulation of fear response(GO:1903367) positive regulation of behavioral fear response(GO:2000987) |
| 0.1 |
0.2 |
GO:0016115 |
terpenoid catabolic process(GO:0016115) |
| 0.1 |
0.2 |
GO:0090285 |
negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.1 |
0.1 |
GO:0048818 |
positive regulation of hair follicle maturation(GO:0048818) positive regulation of catagen(GO:0051795) |
| 0.1 |
0.5 |
GO:0016338 |
calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) establishment of skin barrier(GO:0061436) |
| 0.1 |
0.3 |
GO:0034421 |
post-translational protein acetylation(GO:0034421) |
| 0.1 |
0.2 |
GO:0006097 |
glyoxylate cycle(GO:0006097) |
| 0.1 |
0.2 |
GO:0021586 |
pons maturation(GO:0021586) |
| 0.1 |
0.1 |
GO:1990173 |
protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) protein localization to nucleoplasm(GO:1990173) |
| 0.1 |
0.4 |
GO:0036233 |
glycine import(GO:0036233) |
| 0.1 |
0.4 |
GO:0060923 |
cardiac muscle cell fate commitment(GO:0060923) |
| 0.1 |
0.2 |
GO:0072356 |
chromosome passenger complex localization to kinetochore(GO:0072356) |
| 0.1 |
0.4 |
GO:0070458 |
detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 0.1 |
0.9 |
GO:0050765 |
negative regulation of phagocytosis(GO:0050765) |
| 0.1 |
0.3 |
GO:0032423 |
regulation of mismatch repair(GO:0032423) |
| 0.1 |
0.6 |
GO:0071578 |
zinc II ion transmembrane import(GO:0071578) |
| 0.1 |
0.5 |
GO:0006532 |
aspartate biosynthetic process(GO:0006532) |
| 0.1 |
0.9 |
GO:0006516 |
glycoprotein catabolic process(GO:0006516) |
| 0.1 |
0.3 |
GO:2000255 |
negative regulation of male germ cell proliferation(GO:2000255) |
| 0.1 |
0.2 |
GO:0045039 |
protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 |
0.4 |
GO:0071493 |
cellular response to UV-B(GO:0071493) |
| 0.1 |
0.3 |
GO:0060718 |
chorionic trophoblast cell differentiation(GO:0060718) |
| 0.1 |
0.3 |
GO:0043985 |
histone H4-R3 methylation(GO:0043985) |
| 0.1 |
0.3 |
GO:0070327 |
thyroid hormone transport(GO:0070327) |
| 0.1 |
0.2 |
GO:0032788 |
saturated monocarboxylic acid metabolic process(GO:0032788) unsaturated monocarboxylic acid metabolic process(GO:0032789) |
| 0.0 |
0.4 |
GO:0097034 |
mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 |
0.1 |
GO:0070366 |
regulation of hepatocyte differentiation(GO:0070366) |
| 0.0 |
0.2 |
GO:0038108 |
negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.0 |
0.2 |
GO:1903031 |
regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.0 |
0.4 |
GO:0030836 |
positive regulation of actin filament depolymerization(GO:0030836) |
| 0.0 |
0.2 |
GO:1901341 |
activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 |
0.1 |
GO:0046671 |
negative regulation of retinal cell programmed cell death(GO:0046671) |
| 0.0 |
0.4 |
GO:0061158 |
3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.0 |
0.2 |
GO:0034145 |
positive regulation of toll-like receptor 4 signaling pathway(GO:0034145) |
| 0.0 |
0.1 |
GO:0071895 |
odontoblast differentiation(GO:0071895) |
| 0.0 |
0.3 |
GO:0061635 |
regulation of protein complex stability(GO:0061635) |
| 0.0 |
0.1 |
GO:0060455 |
negative regulation of gastric acid secretion(GO:0060455) |
| 0.0 |
0.1 |
GO:0045113 |
regulation of integrin biosynthetic process(GO:0045113) |
| 0.0 |
0.3 |
GO:1904417 |
regulation of xenophagy(GO:1904415) positive regulation of xenophagy(GO:1904417) |
| 0.0 |
0.3 |
GO:0043504 |
mitochondrial DNA repair(GO:0043504) |
| 0.0 |
2.7 |
GO:0042273 |
ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 |
0.2 |
GO:0043951 |
negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 |
0.1 |
GO:0006556 |
S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 |
0.6 |
GO:0000028 |
ribosomal small subunit assembly(GO:0000028) |
| 0.0 |
0.3 |
GO:0002536 |
respiratory burst involved in inflammatory response(GO:0002536) |
| 0.0 |
1.9 |
GO:0032543 |
mitochondrial translation(GO:0032543) |
| 0.0 |
0.5 |
GO:0006958 |
complement activation, classical pathway(GO:0006958) |
| 0.0 |
0.2 |
GO:0007000 |
nucleolus organization(GO:0007000) |
| 0.0 |
0.1 |
GO:0048619 |
embryonic hindgut morphogenesis(GO:0048619) |
| 0.0 |
0.5 |
GO:2000353 |
positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.0 |
0.1 |
GO:2000271 |
positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 |
0.2 |
GO:0010759 |
positive regulation of macrophage chemotaxis(GO:0010759) |
| 0.0 |
0.2 |
GO:1902847 |
macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of tau-protein kinase activity(GO:1902949) |
| 0.0 |
0.3 |
GO:0048012 |
hepatocyte growth factor receptor signaling pathway(GO:0048012) |
| 0.0 |
0.3 |
GO:0043252 |
sodium-independent organic anion transport(GO:0043252) |
| 0.0 |
0.2 |
GO:1902513 |
regulation of organelle transport along microtubule(GO:1902513) |
| 0.0 |
0.4 |
GO:0046688 |
response to copper ion(GO:0046688) |
| 0.0 |
0.2 |
GO:1904491 |
protein localization to ciliary transition zone(GO:1904491) |
| 0.0 |
0.6 |
GO:0033622 |
integrin activation(GO:0033622) |
| 0.0 |
0.1 |
GO:0015888 |
thiamine transport(GO:0015888) |
| 0.0 |
0.5 |
GO:0031115 |
negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 |
1.0 |
GO:1902476 |
chloride transmembrane transport(GO:1902476) |
| 0.0 |
0.2 |
GO:0007253 |
cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 |
0.1 |
GO:0002184 |
cytoplasmic translational termination(GO:0002184) |
| 0.0 |
0.6 |
GO:0006829 |
zinc II ion transport(GO:0006829) |
| 0.0 |
0.1 |
GO:0006167 |
AMP biosynthetic process(GO:0006167) |
| 0.0 |
0.1 |
GO:1904565 |
response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.0 |
0.1 |
GO:0060155 |
platelet dense granule organization(GO:0060155) |
| 0.0 |
0.2 |
GO:0045583 |
regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 0.0 |
0.3 |
GO:0043249 |
erythrocyte maturation(GO:0043249) |
| 0.0 |
0.1 |
GO:0046104 |
thymidine metabolic process(GO:0046104) |
| 0.0 |
0.1 |
GO:0040032 |
post-embryonic body morphogenesis(GO:0040032) |
| 0.0 |
0.2 |
GO:0007042 |
lysosomal lumen acidification(GO:0007042) |
| 0.0 |
0.3 |
GO:0042541 |
hemoglobin biosynthetic process(GO:0042541) |
| 0.0 |
1.0 |
GO:0006414 |
translational elongation(GO:0006414) |
| 0.0 |
0.7 |
GO:0050873 |
brown fat cell differentiation(GO:0050873) |
| 0.0 |
0.5 |
GO:0070200 |
establishment of protein localization to telomere(GO:0070200) |
| 0.0 |
0.1 |
GO:0045409 |
negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.0 |
0.4 |
GO:1903020 |
positive regulation of glycoprotein metabolic process(GO:1903020) |
| 0.0 |
0.1 |
GO:0048227 |
plasma membrane to endosome transport(GO:0048227) |
| 0.0 |
0.2 |
GO:0043206 |
extracellular fibril organization(GO:0043206) |
| 0.0 |
0.3 |
GO:0001516 |
prostaglandin biosynthetic process(GO:0001516) icosanoid biosynthetic process(GO:0046456) prostanoid biosynthetic process(GO:0046457) fatty acid derivative biosynthetic process(GO:1901570) |
| 0.0 |
0.2 |
GO:0042940 |
D-amino acid transport(GO:0042940) |
| 0.0 |
0.3 |
GO:0030574 |
collagen catabolic process(GO:0030574) |
| 0.0 |
0.1 |
GO:0071442 |
positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.0 |
0.3 |
GO:0010388 |
cullin deneddylation(GO:0010388) |
| 0.0 |
0.6 |
GO:0002089 |
lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 |
0.3 |
GO:0035563 |
positive regulation of chromatin binding(GO:0035563) |
| 0.0 |
0.1 |
GO:0032627 |
interleukin-23 production(GO:0032627) regulation of interleukin-23 production(GO:0032667) |
| 0.0 |
0.1 |
GO:0031038 |
meiotic chromosome movement towards spindle pole(GO:0016344) myosin II filament organization(GO:0031038) regulation of myosin II filament organization(GO:0043519) |
| 0.0 |
0.1 |
GO:1903525 |
regulation of membrane tubulation(GO:1903525) |
| 0.0 |
0.3 |
GO:0032981 |
NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 |
0.1 |
GO:0035356 |
positive regulation of sequestering of triglyceride(GO:0010890) cellular triglyceride homeostasis(GO:0035356) |
| 0.0 |
0.1 |
GO:0006995 |
cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 |
0.1 |
GO:0071494 |
cellular response to UV-C(GO:0071494) |
| 0.0 |
0.2 |
GO:0002021 |
response to dietary excess(GO:0002021) |
| 0.0 |
0.1 |
GO:0070589 |
cell wall mannoprotein biosynthetic process(GO:0000032) mannoprotein metabolic process(GO:0006056) mannoprotein biosynthetic process(GO:0006057) cell wall glycoprotein biosynthetic process(GO:0031506) cell wall biogenesis(GO:0042546) cell wall macromolecule metabolic process(GO:0044036) cell wall macromolecule biosynthetic process(GO:0044038) chain elongation of O-linked mannose residue(GO:0044845) cellular component macromolecule biosynthetic process(GO:0070589) cell wall organization or biogenesis(GO:0071554) |
| 0.0 |
0.1 |
GO:0035093 |
spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.0 |
0.1 |
GO:0070345 |
negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 |
0.6 |
GO:0010862 |
positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 |
0.3 |
GO:0033962 |
cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.0 |
0.1 |
GO:0033299 |
secretion of lysosomal enzymes(GO:0033299) |
| 0.0 |
0.1 |
GO:0090557 |
establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 |
0.3 |
GO:0032968 |
positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 |
0.1 |
GO:0071205 |
clustering of voltage-gated potassium channels(GO:0045163) protein localization to juxtaparanode region of axon(GO:0071205) |
| 0.0 |
0.3 |
GO:0007614 |
short-term memory(GO:0007614) |
| 0.0 |
0.2 |
GO:0000012 |
single strand break repair(GO:0000012) |
| 0.0 |
0.1 |
GO:0061087 |
positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.0 |
0.0 |
GO:0042726 |
flavin-containing compound metabolic process(GO:0042726) |
| 0.0 |
0.8 |
GO:0061077 |
chaperone-mediated protein folding(GO:0061077) |
| 0.0 |
0.0 |
GO:0039530 |
MDA-5 signaling pathway(GO:0039530) |
| 0.0 |
0.1 |
GO:0015868 |
purine ribonucleotide transport(GO:0015868) |
| 0.0 |
0.1 |
GO:0034244 |
negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 |
0.5 |
GO:0034314 |
Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 |
0.7 |
GO:0001881 |
receptor recycling(GO:0001881) |
| 0.0 |
0.5 |
GO:0045600 |
positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 |
0.2 |
GO:0006388 |
tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 |
0.2 |
GO:0048240 |
sperm capacitation(GO:0048240) |
| 0.0 |
0.1 |
GO:0008535 |
respiratory chain complex IV assembly(GO:0008535) |
| 0.0 |
0.6 |
GO:0007200 |
phospholipase C-activating G-protein coupled receptor signaling pathway(GO:0007200) |
| 0.0 |
0.2 |
GO:0034389 |
lipid particle organization(GO:0034389) |
| 0.0 |
0.1 |
GO:0032287 |
peripheral nervous system myelin maintenance(GO:0032287) |
| 0.0 |
0.1 |
GO:0014846 |
esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) regulation of prostatic bud formation(GO:0060685) negative regulation of prostatic bud formation(GO:0060686) |
| 0.0 |
0.1 |
GO:0030322 |
stabilization of membrane potential(GO:0030322) |
| 0.0 |
0.2 |
GO:0070208 |
protein heterotrimerization(GO:0070208) |
| 0.0 |
0.3 |
GO:0006767 |
water-soluble vitamin metabolic process(GO:0006767) |
| 0.0 |
0.1 |
GO:0006729 |
tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 |
0.3 |
GO:0008299 |
isoprenoid biosynthetic process(GO:0008299) |
| 0.0 |
0.1 |
GO:0034393 |
positive regulation of smooth muscle cell apoptotic process(GO:0034393) follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.0 |
0.1 |
GO:0010032 |
meiotic chromosome condensation(GO:0010032) |
| 0.0 |
0.1 |
GO:0060294 |
cilium movement involved in cell motility(GO:0060294) |
| 0.0 |
0.3 |
GO:0071480 |
cellular response to gamma radiation(GO:0071480) |
| 0.0 |
0.2 |
GO:0032897 |
negative regulation of viral transcription(GO:0032897) |
| 0.0 |
0.2 |
GO:0050434 |
positive regulation of viral transcription(GO:0050434) |
| 0.0 |
0.4 |
GO:0006749 |
glutathione metabolic process(GO:0006749) |
| 0.0 |
0.0 |
GO:0006668 |
sphinganine-1-phosphate metabolic process(GO:0006668) |
| 0.0 |
0.5 |
GO:0005978 |
glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 |
0.3 |
GO:0006303 |
double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.0 |
0.1 |
GO:0046415 |
urate metabolic process(GO:0046415) |
| 0.0 |
0.2 |
GO:0034063 |
stress granule assembly(GO:0034063) |
| 0.0 |
0.1 |
GO:2000491 |
positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.0 |
0.2 |
GO:0008053 |
mitochondrial fusion(GO:0008053) |
| 0.0 |
0.3 |
GO:0090382 |
phagosome maturation(GO:0090382) |