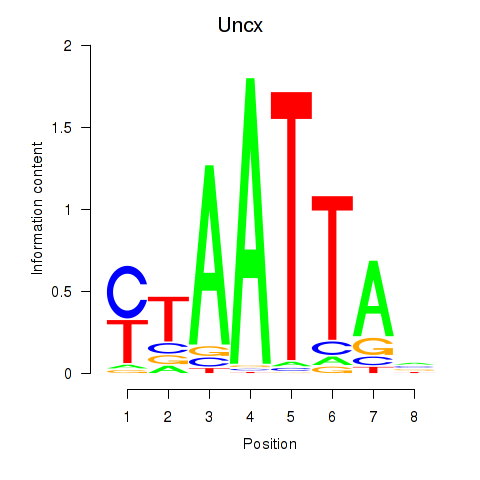
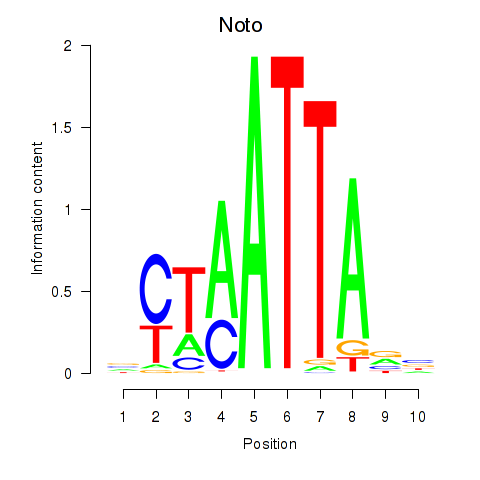
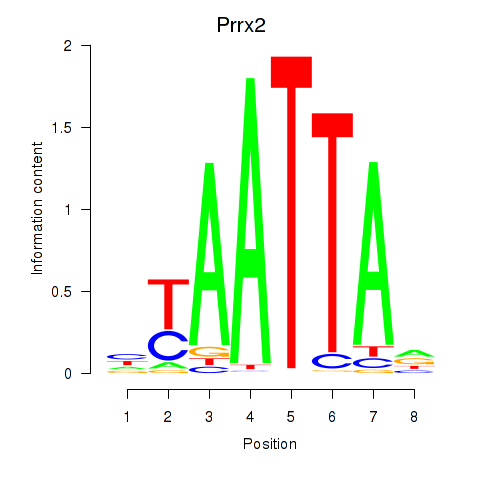
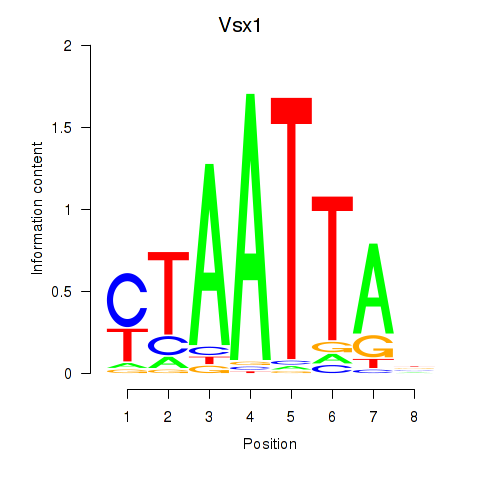
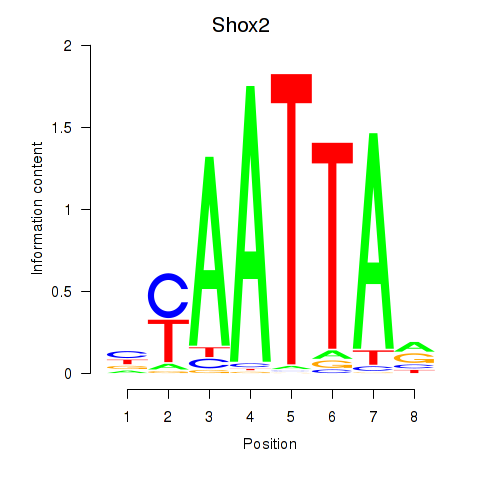
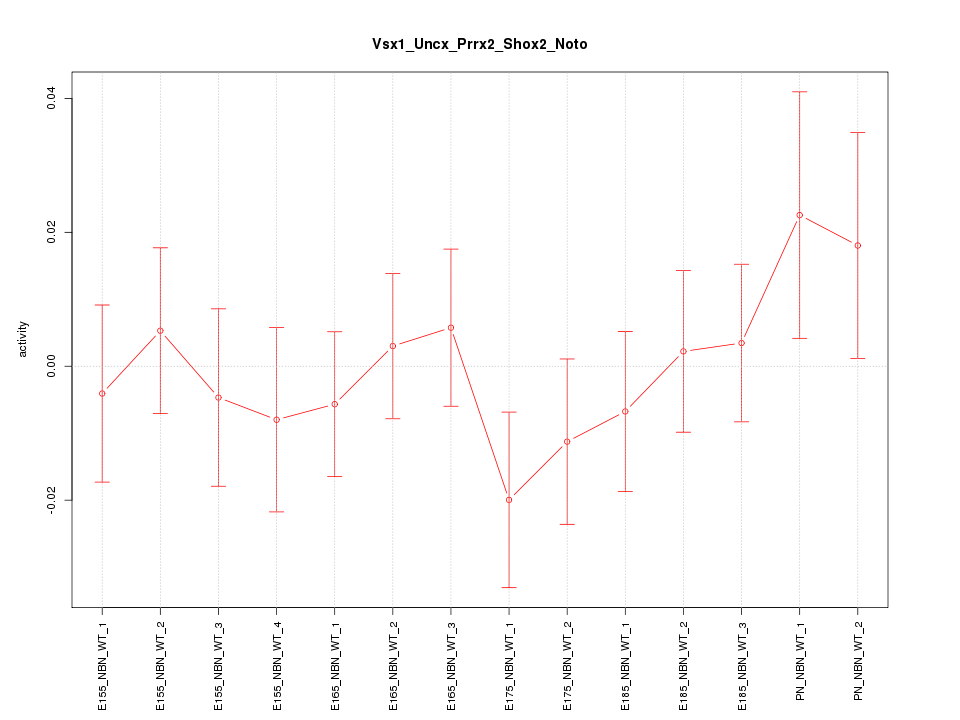
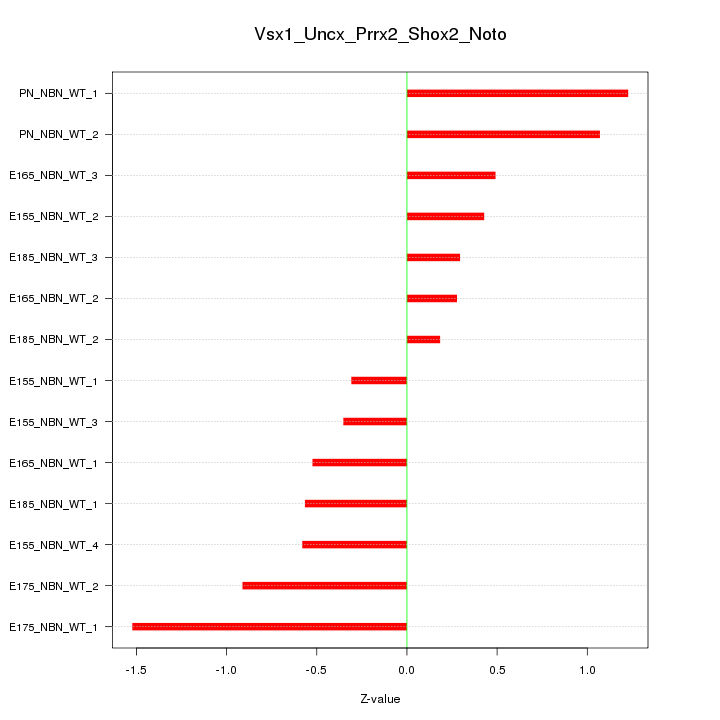
| 1.3 |
4.0 |
GO:0021893 |
cerebral cortex GABAergic interneuron fate commitment(GO:0021893) |
| 1.2 |
3.5 |
GO:0090425 |
hepatocyte cell migration(GO:0002194) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 1.0 |
3.0 |
GO:0002302 |
CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 0.7 |
2.2 |
GO:0003278 |
apoptotic process involved in heart morphogenesis(GO:0003278) regulation of vascular wound healing(GO:0061043) glomerular mesangial cell differentiation(GO:0072008) glomerular mesangial cell development(GO:0072144) |
| 0.6 |
2.5 |
GO:0046552 |
eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.4 |
1.2 |
GO:0008228 |
opsonization(GO:0008228) modulation of molecular function in other organism(GO:0044359) negative regulation of molecular function in other organism(GO:0044362) negative regulation of molecular function in other organism involved in symbiotic interaction(GO:0052204) modulation of molecular function in other organism involved in symbiotic interaction(GO:0052205) negative regulation by host of symbiont molecular function(GO:0052405) modification by host of symbiont molecular function(GO:0052428) |
| 0.4 |
1.1 |
GO:0048389 |
BMP signaling pathway involved in heart induction(GO:0003130) endodermal-mesodermal cell signaling(GO:0003133) endodermal-mesodermal cell signaling involved in heart induction(GO:0003134) intermediate mesoderm development(GO:0048389) regulation of transcription from RNA polymerase II promoter involved in kidney development(GO:0060994) pattern specification involved in mesonephros development(GO:0061227) regulation of hepatocyte differentiation(GO:0070366) comma-shaped body morphogenesis(GO:0072049) S-shaped body morphogenesis(GO:0072050) anterior/posterior pattern specification involved in kidney development(GO:0072098) negative regulation of glomerular mesangial cell proliferation(GO:0072125) ureter urothelium development(GO:0072190) negative regulation of glomerulus development(GO:0090194) regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.3 |
1.5 |
GO:0038032 |
termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.3 |
0.8 |
GO:0015889 |
cobalamin transport(GO:0015889) |
| 0.2 |
1.3 |
GO:0006570 |
tyrosine metabolic process(GO:0006570) |
| 0.2 |
0.6 |
GO:0060166 |
olfactory pit development(GO:0060166) |
| 0.2 |
1.7 |
GO:0043129 |
surfactant homeostasis(GO:0043129) |
| 0.2 |
0.5 |
GO:0035633 |
maintenance of blood-brain barrier(GO:0035633) |
| 0.2 |
2.0 |
GO:0021796 |
cerebral cortex regionalization(GO:0021796) |
| 0.2 |
0.8 |
GO:2000987 |
positive regulation of fear response(GO:1903367) positive regulation of behavioral fear response(GO:2000987) |
| 0.2 |
0.6 |
GO:0003310 |
pancreatic A cell differentiation(GO:0003310) |
| 0.1 |
0.4 |
GO:2000977 |
regulation of forebrain neuron differentiation(GO:2000977) |
| 0.1 |
0.4 |
GO:0019343 |
cysteine biosynthetic process via cystathionine(GO:0019343) cysteine biosynthetic process(GO:0019344) |
| 0.1 |
0.4 |
GO:2000852 |
regulation of corticosterone secretion(GO:2000852) |
| 0.1 |
1.2 |
GO:2000671 |
regulation of motor neuron apoptotic process(GO:2000671) |
| 0.1 |
0.3 |
GO:0061743 |
motor learning(GO:0061743) |
| 0.1 |
0.8 |
GO:0090336 |
positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.1 |
0.7 |
GO:1900194 |
negative regulation of oocyte maturation(GO:1900194) |
| 0.1 |
0.6 |
GO:0043415 |
positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.1 |
0.7 |
GO:0051611 |
negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.1 |
0.8 |
GO:0007195 |
adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.1 |
0.5 |
GO:0007223 |
Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.1 |
0.3 |
GO:0090526 |
response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.1 |
0.4 |
GO:0021780 |
oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.1 |
1.5 |
GO:0006953 |
acute-phase response(GO:0006953) |
| 0.1 |
2.5 |
GO:0007638 |
mechanosensory behavior(GO:0007638) |
| 0.1 |
1.8 |
GO:0033194 |
response to hydroperoxide(GO:0033194) |
| 0.1 |
0.5 |
GO:1901162 |
serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.1 |
0.6 |
GO:0007406 |
negative regulation of neuroblast proliferation(GO:0007406) |
| 0.1 |
0.3 |
GO:0034454 |
microtubule anchoring at centrosome(GO:0034454) |
| 0.1 |
1.4 |
GO:0016339 |
calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 |
0.5 |
GO:0006123 |
mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 |
0.3 |
GO:0072344 |
rescue of stalled ribosome(GO:0072344) |
| 0.1 |
2.4 |
GO:0060074 |
synapse maturation(GO:0060074) |
| 0.1 |
0.3 |
GO:1904565 |
response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.1 |
0.2 |
GO:0030210 |
heparin biosynthetic process(GO:0030210) Tie signaling pathway(GO:0048014) |
| 0.1 |
0.3 |
GO:0000707 |
meiotic DNA recombinase assembly(GO:0000707) resolution of recombination intermediates(GO:0071139) |
| 0.1 |
0.4 |
GO:0072513 |
positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.1 |
1.2 |
GO:0001833 |
inner cell mass cell proliferation(GO:0001833) |
| 0.1 |
0.3 |
GO:0021856 |
cerebral cortex tangential migration using cell-axon interactions(GO:0021824) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) facioacoustic ganglion development(GO:1903375) |
| 0.1 |
0.2 |
GO:0060743 |
epithelial cell maturation involved in prostate gland development(GO:0060743) alveolar secondary septum development(GO:0061144) |
| 0.1 |
0.2 |
GO:0090324 |
negative regulation of oxidative phosphorylation(GO:0090324) |
| 0.1 |
0.3 |
GO:0072592 |
regulation of integrin biosynthetic process(GO:0045113) oxygen metabolic process(GO:0072592) |
| 0.1 |
0.2 |
GO:2001013 |
cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.1 |
2.3 |
GO:0032728 |
positive regulation of interferon-beta production(GO:0032728) |
| 0.1 |
0.2 |
GO:0060023 |
soft palate development(GO:0060023) |
| 0.1 |
3.4 |
GO:0022900 |
electron transport chain(GO:0022900) |
| 0.1 |
0.3 |
GO:0048245 |
eosinophil chemotaxis(GO:0048245) |
| 0.1 |
0.3 |
GO:0006564 |
L-serine biosynthetic process(GO:0006564) |
| 0.1 |
0.1 |
GO:1900377 |
negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.1 |
0.3 |
GO:0060539 |
diaphragm development(GO:0060539) |
| 0.1 |
0.1 |
GO:1904956 |
regulation of midbrain dopaminergic neuron differentiation(GO:1904956) |
| 0.1 |
0.7 |
GO:0031017 |
exocrine pancreas development(GO:0031017) |
| 0.1 |
0.5 |
GO:0010587 |
miRNA catabolic process(GO:0010587) |
| 0.1 |
0.4 |
GO:0045602 |
negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.0 |
0.1 |
GO:0009838 |
abscission(GO:0009838) positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.0 |
1.0 |
GO:0045662 |
negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 |
0.6 |
GO:0071803 |
positive regulation of podosome assembly(GO:0071803) |
| 0.0 |
0.6 |
GO:0050860 |
negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 |
0.5 |
GO:0021521 |
ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.0 |
0.1 |
GO:0010835 |
regulation of protein ADP-ribosylation(GO:0010835) |
| 0.0 |
0.2 |
GO:0051697 |
protein delipidation(GO:0051697) |
| 0.0 |
0.3 |
GO:0060355 |
positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.0 |
0.3 |
GO:0021978 |
telencephalon regionalization(GO:0021978) |
| 0.0 |
0.3 |
GO:0016584 |
nucleosome positioning(GO:0016584) |
| 0.0 |
1.3 |
GO:0031365 |
N-terminal protein amino acid modification(GO:0031365) |
| 0.0 |
0.2 |
GO:0042795 |
snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 |
0.1 |
GO:0001661 |
conditioned taste aversion(GO:0001661) |
| 0.0 |
0.7 |
GO:0002089 |
lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 |
0.1 |
GO:0002326 |
B cell lineage commitment(GO:0002326) |
| 0.0 |
1.3 |
GO:0030901 |
midbrain development(GO:0030901) |
| 0.0 |
0.2 |
GO:0052695 |
cellular glucuronidation(GO:0052695) |
| 0.0 |
0.1 |
GO:0006432 |
phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 |
0.2 |
GO:0070447 |
positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 0.0 |
0.3 |
GO:0060134 |
prepulse inhibition(GO:0060134) |
| 0.0 |
0.3 |
GO:0051764 |
actin crosslink formation(GO:0051764) |
| 0.0 |
0.1 |
GO:0034285 |
response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.0 |
0.4 |
GO:0015937 |
coenzyme A biosynthetic process(GO:0015937) |
| 0.0 |
0.1 |
GO:0010871 |
negative regulation of receptor biosynthetic process(GO:0010871) |
| 0.0 |
2.6 |
GO:0030168 |
platelet activation(GO:0030168) |
| 0.0 |
0.8 |
GO:0007608 |
sensory perception of smell(GO:0007608) |
| 0.0 |
0.1 |
GO:0070562 |
regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 |
0.2 |
GO:0009249 |
protein lipoylation(GO:0009249) |
| 0.0 |
0.2 |
GO:0006003 |
fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 |
0.3 |
GO:0019985 |
translesion synthesis(GO:0019985) |
| 0.0 |
0.2 |
GO:0050957 |
equilibrioception(GO:0050957) |
| 0.0 |
1.1 |
GO:0071300 |
cellular response to retinoic acid(GO:0071300) |
| 0.0 |
0.4 |
GO:0032148 |
activation of protein kinase B activity(GO:0032148) |
| 0.0 |
0.7 |
GO:0006879 |
cellular iron ion homeostasis(GO:0006879) |
| 0.0 |
0.0 |
GO:0042048 |
olfactory behavior(GO:0042048) |
| 0.0 |
0.4 |
GO:0055026 |
negative regulation of cardiac muscle tissue development(GO:0055026) |
| 0.0 |
0.2 |
GO:0010908 |
regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) myoblast fate commitment(GO:0048625) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 |
2.1 |
GO:0019882 |
antigen processing and presentation(GO:0019882) |
| 0.0 |
0.2 |
GO:0043922 |
negative regulation by host of viral transcription(GO:0043922) |
| 0.0 |
0.2 |
GO:0032225 |
regulation of synaptic transmission, dopaminergic(GO:0032225) |
| 0.0 |
0.3 |
GO:0016973 |
poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 |
0.1 |
GO:0015712 |
hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) phosphate ion transmembrane transport(GO:0035435) |
| 0.0 |
0.3 |
GO:0007530 |
sex determination(GO:0007530) |
| 0.0 |
0.2 |
GO:0045579 |
positive regulation of B cell differentiation(GO:0045579) |
| 0.0 |
0.2 |
GO:1903504 |
regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 |
0.4 |
GO:0071108 |
protein K48-linked deubiquitination(GO:0071108) |
| 0.0 |
0.6 |
GO:0051693 |
actin filament capping(GO:0051693) |
| 0.0 |
0.3 |
GO:0080111 |
DNA demethylation(GO:0080111) |
| 0.0 |
0.6 |
GO:0007026 |
negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 |
0.1 |
GO:0045187 |
regulation of circadian sleep/wake cycle(GO:0042749) regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.0 |
0.1 |
GO:0010359 |
regulation of anion channel activity(GO:0010359) |
| 0.0 |
1.3 |
GO:0010811 |
positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 |
0.3 |
GO:0045879 |
negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.0 |
0.1 |
GO:0034047 |
regulation of protein phosphatase type 2A activity(GO:0034047) |
| 0.0 |
0.1 |
GO:0035282 |
segmentation(GO:0035282) |
| 0.0 |
0.0 |
GO:0042420 |
dopamine catabolic process(GO:0042420) |
| 0.0 |
0.2 |
GO:0030497 |
fatty acid elongation(GO:0030497) |
| 0.0 |
0.1 |
GO:0032496 |
response to lipopolysaccharide(GO:0032496) |
| 0.0 |
0.0 |
GO:0010172 |
embryonic body morphogenesis(GO:0010172) |
| 0.0 |
0.1 |
GO:0032049 |
cardiolipin biosynthetic process(GO:0032049) |
| 0.0 |
0.4 |
GO:0048791 |
calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 |
0.3 |
GO:0034723 |
DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 |
0.3 |
GO:1904030 |
negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.0 |
0.2 |
GO:0003301 |
physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 |
0.2 |
GO:0097178 |
ruffle assembly(GO:0097178) |
| 0.0 |
0.1 |
GO:0002361 |
CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |