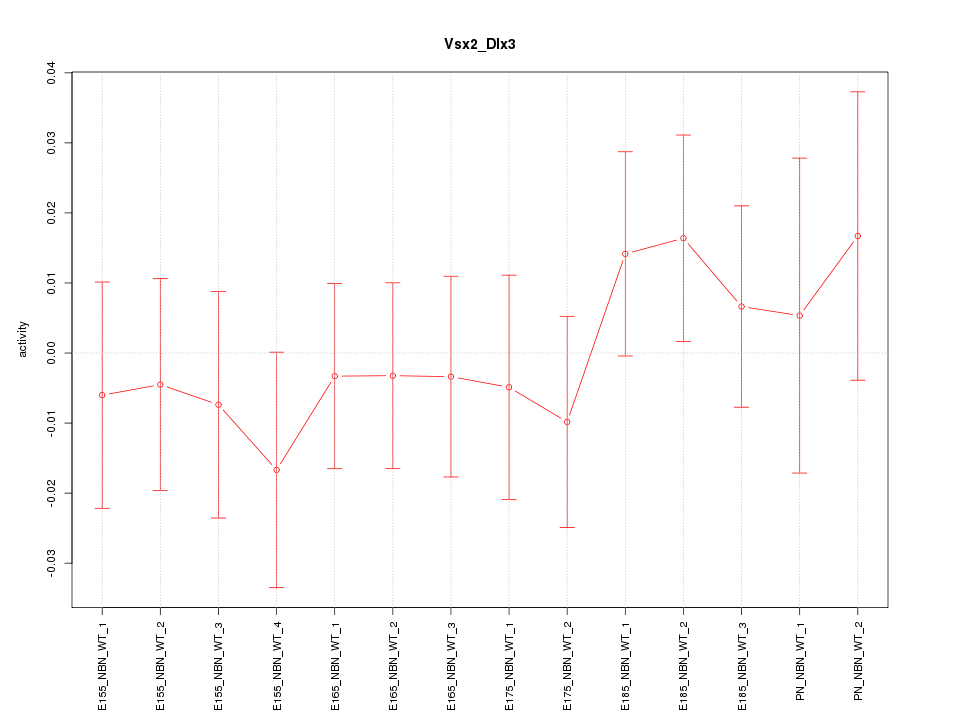
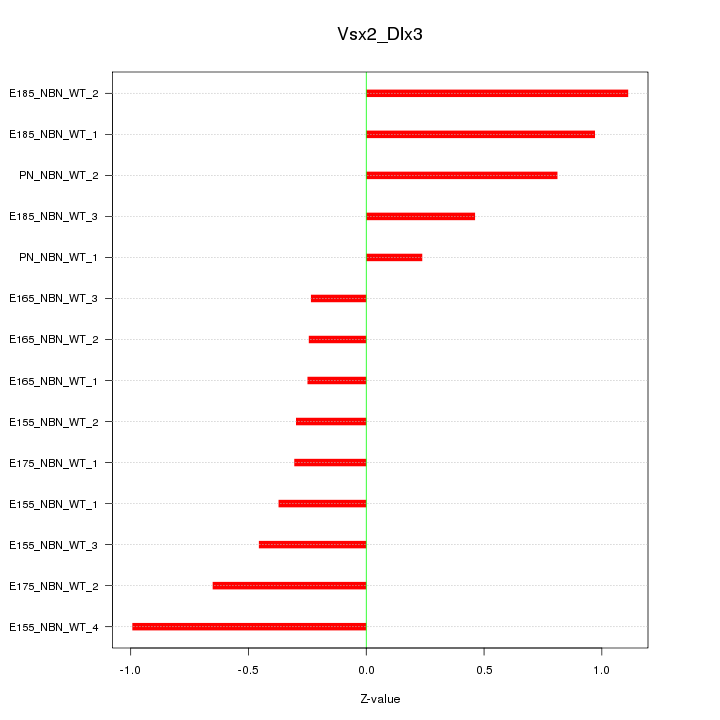
Motif ID: Vsx2_Dlx3
Z-value: 0.611
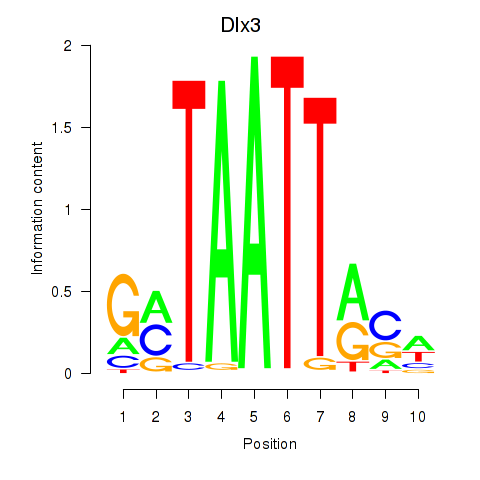
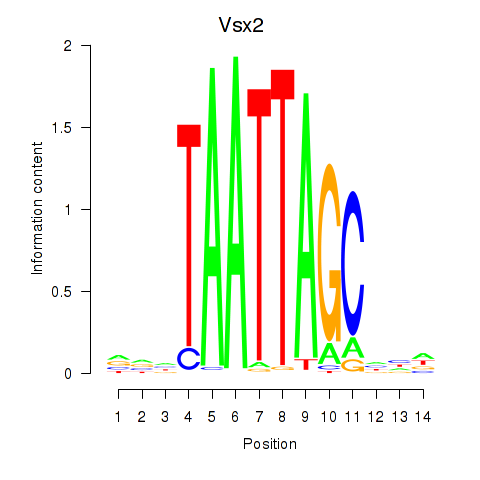
Transcription factors associated with Vsx2_Dlx3:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Dlx3 | ENSMUSG00000001510.7 | Dlx3 |
| Vsx2 | ENSMUSG00000021239.6 | Vsx2 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Vsx2 | mm10_v2_chr12_+_84569762_84569840 | 0.29 | 3.2e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0021893 | cerebral cortex GABAergic interneuron fate commitment(GO:0021893) |
| 0.5 | 1.6 | GO:0090425 | hepatocyte cell migration(GO:0002194) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 0.3 | 1.6 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.3 | 1.0 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.2 | 0.5 | GO:0035633 | maintenance of blood-brain barrier(GO:0035633) |
| 0.1 | 0.6 | GO:1900272 | negative regulation of long-term synaptic potentiation(GO:1900272) |
| 0.1 | 0.8 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.1 | 0.2 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.1 | 0.3 | GO:0090526 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.1 | 0.8 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 0.4 | GO:0046552 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.1 | 0.5 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.1 | 0.9 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.1 | 0.8 | GO:0043129 | surfactant homeostasis(GO:0043129) |
| 0.1 | 1.3 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.1 | 1.4 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.1 | 0.2 | GO:0072204 | cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 0.1 | 0.5 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.1 | 2.0 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.1 | 2.3 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.1 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.0 | 0.4 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.3 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.7 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.5 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.3 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.2 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.6 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.3 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.0 | 0.5 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 0.3 | GO:0031017 | exocrine pancreas development(GO:0031017) |
| 0.0 | 0.3 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.1 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 0.3 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 1.1 | GO:0022900 | electron transport chain(GO:0022900) |
| 0.0 | 0.0 | GO:0002014 | vasoconstriction of artery involved in ischemic response to lowering of systemic arterial blood pressure(GO:0002014) |
| 0.0 | 0.1 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.0 | 0.1 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 0.4 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.4 | GO:0031365 | N-terminal protein amino acid modification(GO:0031365) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) insulin-like growth factor ternary complex(GO:0042567) |
| 0.1 | 0.5 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.1 | 0.4 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.0 | 0.6 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.3 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.3 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 0.5 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 0.9 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.3 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.0 | 0.1 | GO:0030062 | mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.0 | 1.6 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.6 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 1.1 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.3 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.0 | 1.1 | GO:0070469 | respiratory chain(GO:0070469) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 0.2 | 1.6 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.1 | 0.6 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.1 | 0.4 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 0.5 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 0.8 | GO:0005451 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 0.5 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.1 | 0.3 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.1 | 0.5 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 1.1 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 0.3 | GO:0070728 | leucine binding(GO:0070728) |
| 0.1 | 0.8 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 1.1 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.3 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.2 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.5 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.6 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.6 | GO:0050811 | GABA-A receptor activity(GO:0004890) GABA receptor binding(GO:0050811) |
| 0.0 | 0.1 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.0 | 0.3 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 1.3 | GO:0008528 | G-protein coupled peptide receptor activity(GO:0008528) |
| 0.0 | 1.0 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 1.2 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.2 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.5 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 2.2 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 3.5 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 0.3 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.5 | GO:0004601 | peroxidase activity(GO:0004601) |