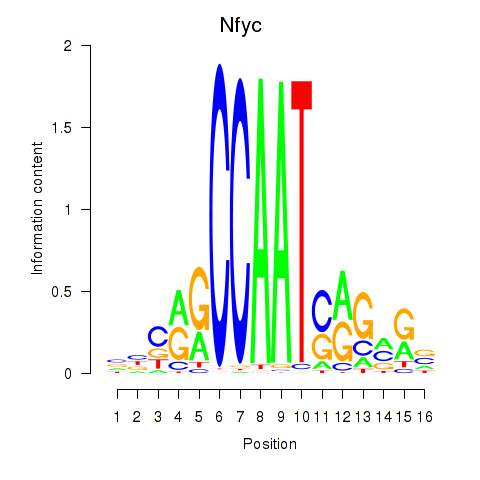
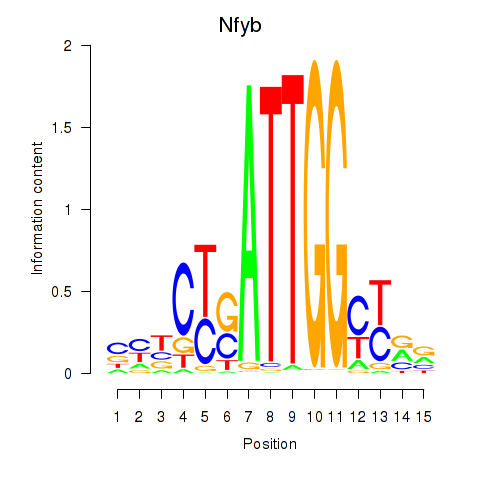
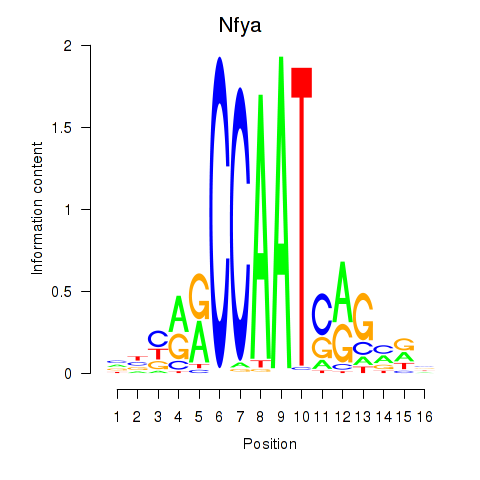
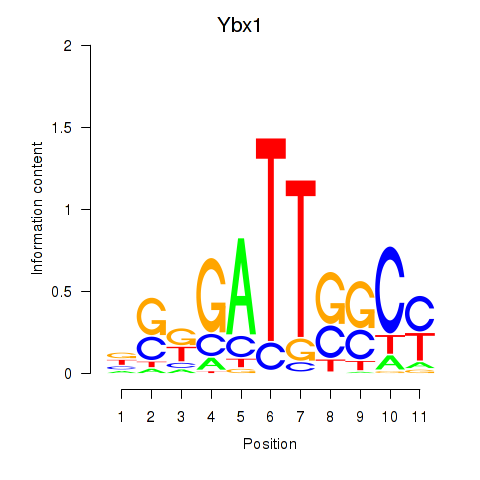
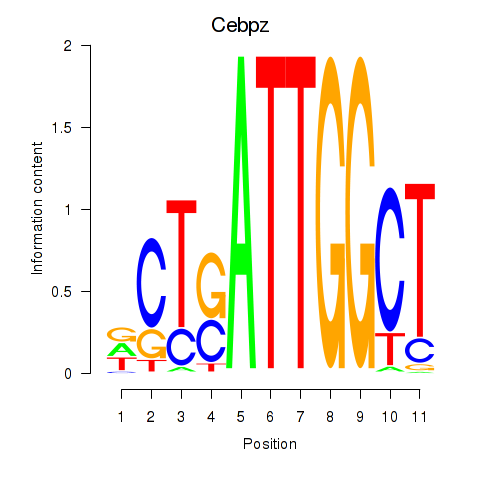
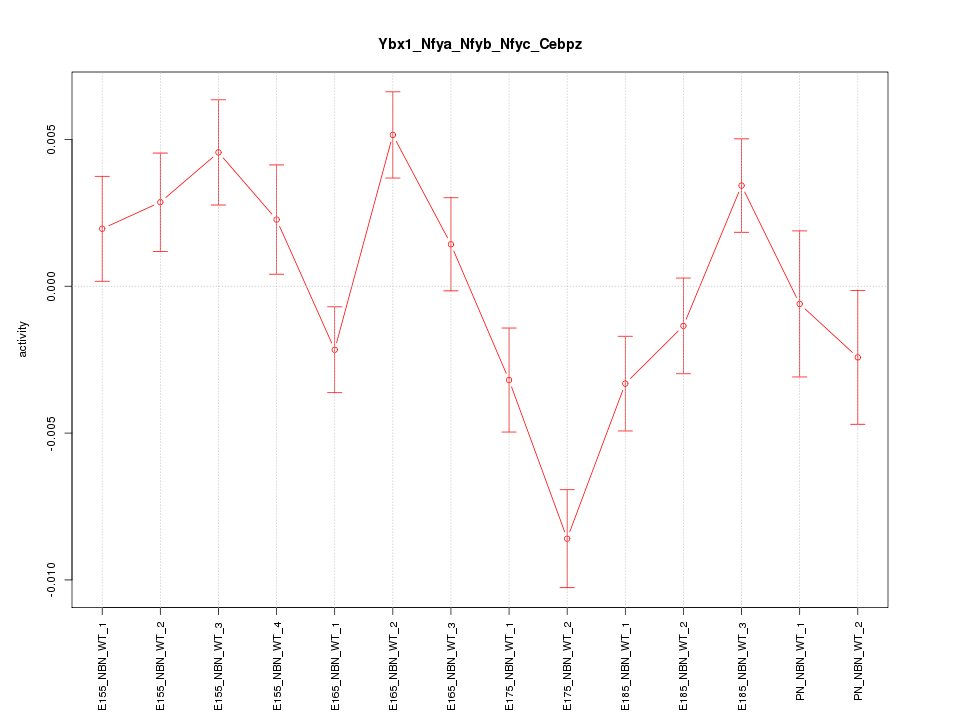
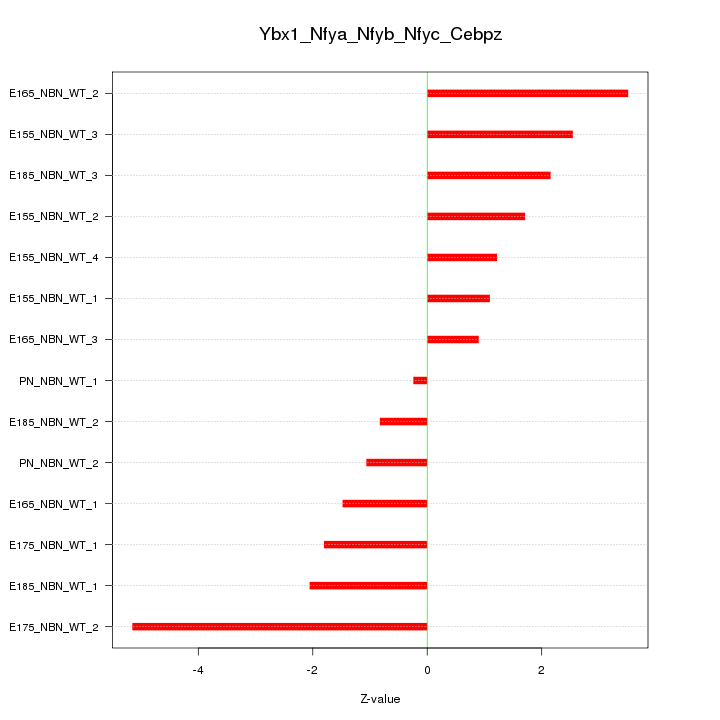
| 2.0 |
5.9 |
GO:0002302 |
CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 0.8 |
3.0 |
GO:0061055 |
myotome development(GO:0061055) |
| 0.6 |
0.6 |
GO:0021957 |
corticospinal tract morphogenesis(GO:0021957) |
| 0.5 |
1.6 |
GO:0002669 |
positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.5 |
1.5 |
GO:0007621 |
negative regulation of female receptivity(GO:0007621) |
| 0.4 |
1.3 |
GO:0061588 |
calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylserine scrambling(GO:0061589) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.4 |
0.8 |
GO:0035910 |
ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 0.4 |
1.2 |
GO:0019401 |
alditol biosynthetic process(GO:0019401) |
| 0.4 |
2.7 |
GO:0031536 |
positive regulation of exit from mitosis(GO:0031536) |
| 0.4 |
1.1 |
GO:0007290 |
spermatid nucleus elongation(GO:0007290) |
| 0.4 |
0.7 |
GO:1900126 |
negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.4 |
1.8 |
GO:0003199 |
endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.4 |
1.1 |
GO:2000297 |
negative regulation of synapse maturation(GO:2000297) |
| 0.4 |
0.7 |
GO:0014735 |
regulation of muscle atrophy(GO:0014735) |
| 0.3 |
1.0 |
GO:0042663 |
regulation of endodermal cell fate specification(GO:0042663) |
| 0.3 |
1.0 |
GO:0006269 |
DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.3 |
1.0 |
GO:1900060 |
negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.3 |
1.3 |
GO:2000427 |
positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.3 |
1.0 |
GO:0030862 |
neuroblast division in subventricular zone(GO:0021849) regulation of polarized epithelial cell differentiation(GO:0030860) positive regulation of polarized epithelial cell differentiation(GO:0030862) |
| 0.3 |
1.3 |
GO:0035795 |
negative regulation of mitochondrial membrane permeability(GO:0035795) |
| 0.3 |
1.3 |
GO:0097393 |
post-embryonic appendage morphogenesis(GO:0035120) post-embryonic limb morphogenesis(GO:0035127) post-embryonic forelimb morphogenesis(GO:0035128) telomeric repeat-containing RNA transcription(GO:0097393) telomeric repeat-containing RNA transcription from RNA pol II promoter(GO:0097394) regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901580) negative regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901581) positive regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901582) |
| 0.3 |
0.9 |
GO:1903351 |
response to dopamine(GO:1903350) cellular response to dopamine(GO:1903351) regulation of sodium:potassium-exchanging ATPase activity(GO:1903406) |
| 0.3 |
0.9 |
GO:0030327 |
prenylated protein catabolic process(GO:0030327) |
| 0.3 |
1.7 |
GO:0060022 |
hard palate development(GO:0060022) |
| 0.3 |
0.8 |
GO:0035880 |
embryonic nail plate morphogenesis(GO:0035880) |
| 0.2 |
0.7 |
GO:0042732 |
D-xylose metabolic process(GO:0042732) |
| 0.2 |
2.3 |
GO:1900112 |
regulation of histone H3-K9 trimethylation(GO:1900112) negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.2 |
0.8 |
GO:0097167 |
circadian regulation of translation(GO:0097167) |
| 0.2 |
0.6 |
GO:2000744 |
anterior head development(GO:0097065) regulation of anterior head development(GO:2000742) positive regulation of anterior head development(GO:2000744) |
| 0.2 |
0.6 |
GO:0006931 |
substrate-dependent cell migration, cell attachment to substrate(GO:0006931) calcium-independent cell-matrix adhesion(GO:0007161) |
| 0.2 |
0.6 |
GO:0002014 |
vasoconstriction of artery involved in ischemic response to lowering of systemic arterial blood pressure(GO:0002014) |
| 0.2 |
0.6 |
GO:1902605 |
heterotrimeric G-protein complex assembly(GO:1902605) |
| 0.2 |
0.6 |
GO:0050975 |
sensory perception of touch(GO:0050975) |
| 0.2 |
0.6 |
GO:0045819 |
positive regulation of glycogen catabolic process(GO:0045819) |
| 0.2 |
1.8 |
GO:0090166 |
Golgi disassembly(GO:0090166) |
| 0.2 |
0.6 |
GO:1901552 |
positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.2 |
0.4 |
GO:0043490 |
malate-aspartate shuttle(GO:0043490) |
| 0.2 |
1.3 |
GO:0048102 |
autophagic cell death(GO:0048102) |
| 0.2 |
1.6 |
GO:0042095 |
interferon-gamma biosynthetic process(GO:0042095) |
| 0.2 |
0.5 |
GO:1903232 |
melanosome assembly(GO:1903232) |
| 0.2 |
0.5 |
GO:1902174 |
positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.2 |
0.5 |
GO:0006428 |
isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.2 |
0.3 |
GO:0090202 |
transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.2 |
2.7 |
GO:0033234 |
negative regulation of protein sumoylation(GO:0033234) |
| 0.2 |
1.1 |
GO:0035845 |
photoreceptor cell outer segment organization(GO:0035845) |
| 0.2 |
0.5 |
GO:0090403 |
oxidative stress-induced premature senescence(GO:0090403) |
| 0.2 |
0.8 |
GO:0051684 |
maintenance of Golgi location(GO:0051684) |
| 0.1 |
0.4 |
GO:1903054 |
lysosomal membrane organization(GO:0097212) negative regulation of extracellular matrix organization(GO:1903054) |
| 0.1 |
0.4 |
GO:0001193 |
maintenance of transcriptional fidelity during DNA-templated transcription elongation(GO:0001192) maintenance of transcriptional fidelity during DNA-templated transcription elongation from RNA polymerase II promoter(GO:0001193) |
| 0.1 |
0.4 |
GO:0016321 |
female meiosis chromosome segregation(GO:0016321) |
| 0.1 |
0.3 |
GO:1903215 |
negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.1 |
0.9 |
GO:0006621 |
protein retention in ER lumen(GO:0006621) |
| 0.1 |
1.1 |
GO:0006537 |
glutamate biosynthetic process(GO:0006537) |
| 0.1 |
0.5 |
GO:0010510 |
regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.1 |
1.6 |
GO:0034773 |
histone H4-K20 trimethylation(GO:0034773) |
| 0.1 |
0.8 |
GO:0060017 |
parathyroid gland development(GO:0060017) |
| 0.1 |
0.5 |
GO:0001771 |
immunological synapse formation(GO:0001771) |
| 0.1 |
0.3 |
GO:0061198 |
fungiform papilla development(GO:0061196) fungiform papilla morphogenesis(GO:0061197) fungiform papilla formation(GO:0061198) |
| 0.1 |
0.2 |
GO:0035519 |
protein K29-linked ubiquitination(GO:0035519) |
| 0.1 |
0.9 |
GO:0006551 |
leucine metabolic process(GO:0006551) |
| 0.1 |
2.0 |
GO:2000251 |
positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 |
0.5 |
GO:0006933 |
negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.1 |
0.4 |
GO:0045659 |
regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.1 |
1.4 |
GO:0044387 |
negative regulation of protein kinase activity by regulation of protein phosphorylation(GO:0044387) |
| 0.1 |
0.4 |
GO:0000087 |
mitotic M phase(GO:0000087) |
| 0.1 |
0.5 |
GO:0043973 |
histone H3-K4 acetylation(GO:0043973) |
| 0.1 |
0.5 |
GO:0090365 |
regulation of mRNA modification(GO:0090365) |
| 0.1 |
0.4 |
GO:1901675 |
response to methylglyoxal(GO:0051595) negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.1 |
0.4 |
GO:0018199 |
peptidyl-glutamine modification(GO:0018199) |
| 0.1 |
1.1 |
GO:0010826 |
negative regulation of centrosome duplication(GO:0010826) negative regulation of centrosome cycle(GO:0046606) |
| 0.1 |
1.4 |
GO:0007100 |
mitotic centrosome separation(GO:0007100) |
| 0.1 |
0.8 |
GO:0001887 |
selenium compound metabolic process(GO:0001887) |
| 0.1 |
0.6 |
GO:1904721 |
regulation of mRNA cleavage(GO:0031437) negative regulation of mRNA cleavage(GO:0031438) negative regulation of immunoglobulin secretion(GO:0051025) regulation of endoribonuclease activity(GO:0060699) negative regulation of ribonuclease activity(GO:0060701) negative regulation of endoribonuclease activity(GO:0060702) regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904720) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.1 |
0.6 |
GO:0072429 |
response to cell cycle checkpoint signaling(GO:0072396) response to DNA integrity checkpoint signaling(GO:0072402) response to DNA damage checkpoint signaling(GO:0072423) response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.1 |
0.5 |
GO:0035093 |
spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.1 |
0.3 |
GO:0000965 |
mitochondrial RNA 3'-end processing(GO:0000965) |
| 0.1 |
0.1 |
GO:0035984 |
response to trichostatin A(GO:0035983) cellular response to trichostatin A(GO:0035984) |
| 0.1 |
1.0 |
GO:0018230 |
peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 |
1.4 |
GO:0007141 |
male meiosis I(GO:0007141) |
| 0.1 |
0.4 |
GO:0046836 |
glycolipid transport(GO:0046836) |
| 0.1 |
2.1 |
GO:0016226 |
iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.1 |
1.1 |
GO:0036499 |
PERK-mediated unfolded protein response(GO:0036499) |
| 0.1 |
0.3 |
GO:0042822 |
vitamin B6 metabolic process(GO:0042816) pyridoxal phosphate metabolic process(GO:0042822) pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.1 |
0.3 |
GO:0060050 |
positive regulation of protein glycosylation(GO:0060050) |
| 0.1 |
0.3 |
GO:0060672 |
epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.1 |
0.4 |
GO:0071609 |
chemokine (C-C motif) ligand 5 production(GO:0071609) |
| 0.1 |
0.3 |
GO:0035604 |
fibroblast growth factor receptor signaling pathway involved in negative regulation of apoptotic process in bone marrow(GO:0035602) fibroblast growth factor receptor signaling pathway involved in hemopoiesis(GO:0035603) fibroblast growth factor receptor signaling pathway involved in positive regulation of cell proliferation in bone marrow(GO:0035604) coronal suture morphogenesis(GO:0060365) squamous basal epithelial stem cell differentiation involved in prostate gland acinus development(GO:0060529) |
| 0.1 |
0.2 |
GO:0045409 |
negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.1 |
0.5 |
GO:1900147 |
Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.1 |
0.2 |
GO:0010847 |
regulation of chromatin assembly(GO:0010847) |
| 0.1 |
0.3 |
GO:0048819 |
regulation of hair follicle maturation(GO:0048819) regulation of catagen(GO:0051794) |
| 0.1 |
0.4 |
GO:0034239 |
regulation of macrophage fusion(GO:0034239) |
| 0.1 |
0.4 |
GO:0010808 |
positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.1 |
0.3 |
GO:0019482 |
beta-alanine metabolic process(GO:0019482) |
| 0.1 |
0.8 |
GO:0006013 |
mannose metabolic process(GO:0006013) |
| 0.1 |
0.3 |
GO:0060155 |
platelet dense granule organization(GO:0060155) |
| 0.1 |
0.7 |
GO:0006477 |
protein sulfation(GO:0006477) |
| 0.1 |
0.4 |
GO:0000733 |
DNA strand renaturation(GO:0000733) |
| 0.1 |
4.0 |
GO:0048821 |
erythrocyte development(GO:0048821) |
| 0.1 |
0.2 |
GO:1903377 |
negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.1 |
1.3 |
GO:0034501 |
protein localization to kinetochore(GO:0034501) |
| 0.1 |
0.3 |
GO:2000620 |
positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.1 |
2.8 |
GO:0034723 |
DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 |
0.1 |
GO:0006404 |
RNA import into nucleus(GO:0006404) |
| 0.1 |
0.7 |
GO:0051574 |
positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.1 |
0.3 |
GO:0007023 |
post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.1 |
0.1 |
GO:0051026 |
chiasma assembly(GO:0051026) |
| 0.1 |
0.5 |
GO:0030644 |
cellular chloride ion homeostasis(GO:0030644) |
| 0.1 |
0.6 |
GO:2000653 |
regulation of genetic imprinting(GO:2000653) |
| 0.1 |
0.1 |
GO:0006106 |
fumarate metabolic process(GO:0006106) |
| 0.1 |
0.2 |
GO:0032650 |
regulation of interleukin-1 alpha production(GO:0032650) positive regulation of interleukin-1 alpha production(GO:0032730) |
| 0.1 |
0.4 |
GO:0006627 |
protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.1 |
0.4 |
GO:0050862 |
positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.1 |
1.1 |
GO:0043968 |
histone H2A acetylation(GO:0043968) |
| 0.1 |
0.6 |
GO:1902177 |
positive regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902177) |
| 0.1 |
0.6 |
GO:2000074 |
regulation of type B pancreatic cell development(GO:2000074) |
| 0.1 |
0.4 |
GO:0090292 |
nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.1 |
0.2 |
GO:2000832 |
protein-chromophore linkage(GO:0018298) negative regulation of steroid hormone secretion(GO:2000832) negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.1 |
7.9 |
GO:0006342 |
chromatin silencing(GO:0006342) |
| 0.1 |
0.7 |
GO:0035881 |
amacrine cell differentiation(GO:0035881) |
| 0.1 |
0.3 |
GO:0072344 |
rescue of stalled ribosome(GO:0072344) |
| 0.1 |
0.2 |
GO:0042536 |
negative regulation of tumor necrosis factor biosynthetic process(GO:0042536) |
| 0.1 |
0.2 |
GO:0035414 |
negative regulation of catenin import into nucleus(GO:0035414) |
| 0.1 |
0.1 |
GO:1901668 |
regulation of superoxide dismutase activity(GO:1901668) |
| 0.1 |
0.8 |
GO:0097034 |
mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.1 |
0.3 |
GO:0051987 |
positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) |
| 0.1 |
0.3 |
GO:0052490 |
suppression by virus of host apoptotic process(GO:0019050) negative regulation by symbiont of host apoptotic process(GO:0033668) modulation by virus of host apoptotic process(GO:0039526) response to cycloheximide(GO:0046898) negative regulation by symbiont of host programmed cell death(GO:0052041) negative regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052490) |
| 0.1 |
0.4 |
GO:0006957 |
complement activation, alternative pathway(GO:0006957) |
| 0.1 |
0.1 |
GO:0042359 |
vitamin D metabolic process(GO:0042359) |
| 0.1 |
0.5 |
GO:0051004 |
regulation of lipoprotein lipase activity(GO:0051004) |
| 0.1 |
0.3 |
GO:0071139 |
resolution of recombination intermediates(GO:0071139) |
| 0.1 |
0.1 |
GO:1990705 |
cholangiocyte proliferation(GO:1990705) |
| 0.1 |
0.4 |
GO:0048631 |
regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.1 |
0.3 |
GO:0036337 |
Fas signaling pathway(GO:0036337) |
| 0.1 |
1.3 |
GO:0099517 |
anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.1 |
0.2 |
GO:0007210 |
serotonin receptor signaling pathway(GO:0007210) |
| 0.1 |
0.8 |
GO:0002227 |
innate immune response in mucosa(GO:0002227) |
| 0.1 |
0.5 |
GO:2000628 |
regulation of miRNA metabolic process(GO:2000628) |
| 0.1 |
1.1 |
GO:0019835 |
cytolysis(GO:0019835) |
| 0.1 |
0.5 |
GO:1904668 |
positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.1 |
0.1 |
GO:0061743 |
motor learning(GO:0061743) |
| 0.1 |
0.1 |
GO:0006867 |
asparagine transport(GO:0006867) glutamine transport(GO:0006868) |
| 0.1 |
2.4 |
GO:0043551 |
regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.1 |
1.1 |
GO:1900745 |
positive regulation of p38MAPK cascade(GO:1900745) |
| 0.1 |
0.1 |
GO:0002636 |
positive regulation of germinal center formation(GO:0002636) |
| 0.1 |
0.3 |
GO:0045903 |
positive regulation of translational fidelity(GO:0045903) |
| 0.1 |
0.2 |
GO:0048162 |
preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.1 |
0.9 |
GO:0043508 |
negative regulation of JUN kinase activity(GO:0043508) |
| 0.1 |
0.8 |
GO:0030150 |
protein import into mitochondrial matrix(GO:0030150) |
| 0.1 |
0.7 |
GO:0070244 |
negative regulation of thymocyte apoptotic process(GO:0070244) |
| 0.1 |
0.9 |
GO:0021684 |
cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.1 |
0.2 |
GO:0019676 |
ammonia assimilation cycle(GO:0019676) |
| 0.1 |
0.3 |
GO:0033136 |
serine phosphorylation of STAT3 protein(GO:0033136) |
| 0.1 |
0.1 |
GO:0061300 |
cerebellum vasculature development(GO:0061300) |
| 0.1 |
0.1 |
GO:0071459 |
protein localization to chromosome, centromeric region(GO:0071459) |
| 0.1 |
0.1 |
GO:0002339 |
B cell selection(GO:0002339) |
| 0.1 |
0.4 |
GO:0006287 |
base-excision repair, gap-filling(GO:0006287) |
| 0.1 |
0.2 |
GO:0003278 |
apoptotic process involved in heart morphogenesis(GO:0003278) |
| 0.1 |
0.3 |
GO:0071896 |
protein localization to adherens junction(GO:0071896) |
| 0.1 |
0.3 |
GO:0046654 |
tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.1 |
1.2 |
GO:0051443 |
positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.1 |
0.1 |
GO:0001656 |
metanephros development(GO:0001656) |
| 0.1 |
0.2 |
GO:0072531 |
pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.1 |
0.4 |
GO:0060536 |
cartilage morphogenesis(GO:0060536) |
| 0.1 |
0.1 |
GO:0043201 |
response to leucine(GO:0043201) cellular response to leucine(GO:0071233) |
| 0.1 |
0.2 |
GO:0002036 |
regulation of L-glutamate transport(GO:0002036) |
| 0.1 |
0.7 |
GO:0015697 |
quaternary ammonium group transport(GO:0015697) |
| 0.1 |
0.2 |
GO:0006546 |
glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.1 |
0.2 |
GO:0032918 |
polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.1 |
0.5 |
GO:1990440 |
positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.1 |
0.2 |
GO:1902416 |
positive regulation of mRNA binding(GO:1902416) positive regulation of RNA binding(GO:1905216) |
| 0.1 |
0.1 |
GO:0009051 |
pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.1 |
0.4 |
GO:0034638 |
phosphatidylcholine catabolic process(GO:0034638) |
| 0.1 |
0.1 |
GO:0035087 |
targeting of mRNA for destruction involved in RNA interference(GO:0030423) siRNA loading onto RISC involved in RNA interference(GO:0035087) |
| 0.1 |
0.2 |
GO:0072369 |
regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 0.1 |
0.2 |
GO:2001168 |
regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.1 |
0.3 |
GO:0098789 |
pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 |
0.2 |
GO:0006566 |
threonine metabolic process(GO:0006566) |
| 0.1 |
2.5 |
GO:0006506 |
GPI anchor biosynthetic process(GO:0006506) |
| 0.1 |
0.2 |
GO:0070589 |
cell wall mannoprotein biosynthetic process(GO:0000032) mannoprotein metabolic process(GO:0006056) mannoprotein biosynthetic process(GO:0006057) cell wall glycoprotein biosynthetic process(GO:0031506) cell wall biogenesis(GO:0042546) cell wall macromolecule metabolic process(GO:0044036) cell wall macromolecule biosynthetic process(GO:0044038) chain elongation of O-linked mannose residue(GO:0044845) cellular component macromolecule biosynthetic process(GO:0070589) cell wall organization or biogenesis(GO:0071554) |
| 0.1 |
0.4 |
GO:0006561 |
proline biosynthetic process(GO:0006561) |
| 0.1 |
1.0 |
GO:0045648 |
positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.1 |
0.5 |
GO:0045116 |
protein neddylation(GO:0045116) |
| 0.1 |
0.2 |
GO:2000612 |
positive regulation of mesenchymal to epithelial transition involved in metanephros morphogenesis(GO:0072108) regulation of mesenchymal cell apoptotic process involved in metanephric nephron morphogenesis(GO:0072304) negative regulation of mesenchymal cell apoptotic process involved in metanephric nephron morphogenesis(GO:0072305) mesenchymal cell apoptotic process involved in metanephros development(GO:1900200) apoptotic process involved in metanephric collecting duct development(GO:1900204) apoptotic process involved in metanephric nephron tubule development(GO:1900205) regulation of mesenchymal cell apoptotic process involved in metanephros development(GO:1900211) negative regulation of mesenchymal cell apoptotic process involved in metanephros development(GO:1900212) regulation of apoptotic process involved in metanephric collecting duct development(GO:1900214) negative regulation of apoptotic process involved in metanephric collecting duct development(GO:1900215) regulation of apoptotic process involved in metanephric nephron tubule development(GO:1900217) negative regulation of apoptotic process involved in metanephric nephron tubule development(GO:1900218) mesenchymal cell apoptotic process involved in metanephric nephron morphogenesis(GO:1901147) regulation of metanephric DCT cell differentiation(GO:2000592) positive regulation of metanephric DCT cell differentiation(GO:2000594) regulation of thyroid hormone generation(GO:2000609) positive regulation of thyroid hormone generation(GO:2000611) regulation of thyroid-stimulating hormone secretion(GO:2000612) |
| 0.1 |
0.2 |
GO:0002326 |
B cell lineage commitment(GO:0002326) |
| 0.1 |
0.5 |
GO:0006301 |
postreplication repair(GO:0006301) |
| 0.1 |
0.3 |
GO:0070934 |
CRD-mediated mRNA stabilization(GO:0070934) |
| 0.1 |
0.3 |
GO:0032298 |
positive regulation of DNA-dependent DNA replication initiation(GO:0032298) |
| 0.1 |
0.4 |
GO:1900262 |
regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.1 |
0.1 |
GO:0006533 |
aspartate catabolic process(GO:0006533) |
| 0.1 |
0.1 |
GO:0007063 |
regulation of sister chromatid cohesion(GO:0007063) |
| 0.1 |
0.4 |
GO:0035542 |
regulation of SNARE complex assembly(GO:0035542) |
| 0.1 |
0.3 |
GO:0070561 |
vitamin D receptor signaling pathway(GO:0070561) |
| 0.1 |
0.1 |
GO:0021759 |
globus pallidus development(GO:0021759) |
| 0.1 |
0.2 |
GO:0035092 |
sperm chromatin condensation(GO:0035092) |
| 0.1 |
0.1 |
GO:0006624 |
vacuolar protein processing(GO:0006624) |
| 0.1 |
0.8 |
GO:0033327 |
Leydig cell differentiation(GO:0033327) |
| 0.0 |
0.4 |
GO:0006449 |
regulation of translational termination(GO:0006449) |
| 0.0 |
0.2 |
GO:0002035 |
brain renin-angiotensin system(GO:0002035) |
| 0.0 |
1.0 |
GO:0045947 |
negative regulation of translational initiation(GO:0045947) |
| 0.0 |
0.2 |
GO:0035509 |
negative regulation of myosin-light-chain-phosphatase activity(GO:0035509) |
| 0.0 |
0.1 |
GO:2000983 |
regulation of ATP citrate synthase activity(GO:2000983) negative regulation of ATP citrate synthase activity(GO:2000984) |
| 0.0 |
0.1 |
GO:0019230 |
proprioception(GO:0019230) |
| 0.0 |
0.3 |
GO:0034587 |
piRNA metabolic process(GO:0034587) |
| 0.0 |
0.1 |
GO:0071374 |
cellular response to parathyroid hormone stimulus(GO:0071374) |
| 0.0 |
0.1 |
GO:0009182 |
purine deoxyribonucleoside diphosphate metabolic process(GO:0009182) dGDP metabolic process(GO:0046066) GDP metabolic process(GO:0046710) |
| 0.0 |
0.3 |
GO:0061668 |
mitochondrial ribosome assembly(GO:0061668) |
| 0.0 |
0.0 |
GO:0034637 |
cellular carbohydrate biosynthetic process(GO:0034637) |
| 0.0 |
0.2 |
GO:1903237 |
negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.0 |
0.6 |
GO:0021796 |
cerebral cortex regionalization(GO:0021796) |
| 0.0 |
0.4 |
GO:0071941 |
nitrogen cycle metabolic process(GO:0071941) |
| 0.0 |
0.5 |
GO:1900037 |
regulation of cellular response to hypoxia(GO:1900037) |
| 0.0 |
0.4 |
GO:0042760 |
very long-chain fatty acid catabolic process(GO:0042760) |
| 0.0 |
0.5 |
GO:2000010 |
positive regulation of protein localization to cell surface(GO:2000010) |
| 0.0 |
0.6 |
GO:2000009 |
negative regulation of protein localization to cell surface(GO:2000009) |
| 0.0 |
0.0 |
GO:0060414 |
aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 |
0.3 |
GO:1903690 |
negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 |
0.3 |
GO:0060028 |
convergent extension involved in axis elongation(GO:0060028) |
| 0.0 |
1.2 |
GO:0015985 |
energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 |
0.5 |
GO:0051457 |
maintenance of protein location in nucleus(GO:0051457) |
| 0.0 |
0.0 |
GO:1902861 |
copper ion import into cell(GO:1902861) |
| 0.0 |
0.0 |
GO:1901979 |
regulation of inward rectifier potassium channel activity(GO:1901979) |
| 0.0 |
0.6 |
GO:0006817 |
phosphate ion transport(GO:0006817) |
| 0.0 |
0.2 |
GO:0034727 |
lysosomal microautophagy(GO:0016237) piecemeal microautophagy of nucleus(GO:0034727) late nucleophagy(GO:0044805) |
| 0.0 |
0.2 |
GO:0045842 |
positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.0 |
0.1 |
GO:2001137 |
positive regulation of endocytic recycling(GO:2001137) |
| 0.0 |
0.2 |
GO:0035166 |
post-embryonic hemopoiesis(GO:0035166) |
| 0.0 |
0.6 |
GO:0021542 |
dentate gyrus development(GO:0021542) |
| 0.0 |
0.6 |
GO:0070935 |
3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 |
0.3 |
GO:0042984 |
amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.0 |
0.1 |
GO:0006419 |
alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 |
0.4 |
GO:1900121 |
negative regulation of receptor binding(GO:1900121) |
| 0.0 |
0.4 |
GO:0042989 |
sequestering of actin monomers(GO:0042989) |
| 0.0 |
0.2 |
GO:0016540 |
protein autoprocessing(GO:0016540) |
| 0.0 |
0.1 |
GO:0006435 |
threonyl-tRNA aminoacylation(GO:0006435) |
| 0.0 |
0.4 |
GO:0070886 |
positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 |
0.4 |
GO:0006107 |
oxaloacetate metabolic process(GO:0006107) |
| 0.0 |
0.2 |
GO:0000480 |
endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 |
0.4 |
GO:0036376 |
sodium ion export from cell(GO:0036376) |
| 0.0 |
0.6 |
GO:0000731 |
DNA synthesis involved in DNA repair(GO:0000731) |
| 0.0 |
0.1 |
GO:1990253 |
cellular response to leucine starvation(GO:1990253) |
| 0.0 |
0.6 |
GO:0006388 |
tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 |
0.5 |
GO:0050910 |
detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.0 |
1.4 |
GO:0045824 |
negative regulation of innate immune response(GO:0045824) |
| 0.0 |
0.1 |
GO:2000393 |
negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.0 |
0.2 |
GO:2000211 |
regulation of glutamate metabolic process(GO:2000211) |
| 0.0 |
0.1 |
GO:0030397 |
membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.0 |
0.1 |
GO:0035672 |
transepithelial chloride transport(GO:0030321) oligopeptide transmembrane transport(GO:0035672) |
| 0.0 |
2.2 |
GO:0015807 |
L-amino acid transport(GO:0015807) |
| 0.0 |
0.1 |
GO:0001978 |
regulation of systemic arterial blood pressure by carotid sinus baroreceptor feedback(GO:0001978) |
| 0.0 |
0.1 |
GO:1904431 |
positive regulation of t-circle formation(GO:1904431) |
| 0.0 |
0.1 |
GO:0031038 |
myosin II filament organization(GO:0031038) regulation of myosin II filament organization(GO:0043519) |
| 0.0 |
0.0 |
GO:0034497 |
protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 |
0.2 |
GO:0032472 |
Golgi calcium ion transport(GO:0032472) |
| 0.0 |
0.2 |
GO:1900451 |
positive regulation of glutamate receptor signaling pathway(GO:1900451) positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 0.0 |
0.1 |
GO:0018103 |
protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 |
0.1 |
GO:0000479 |
endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000479) |
| 0.0 |
0.1 |
GO:0060398 |
regulation of growth hormone receptor signaling pathway(GO:0060398) positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.0 |
0.1 |
GO:0006669 |
sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.0 |
0.3 |
GO:0035608 |
protein deglutamylation(GO:0035608) |
| 0.0 |
0.1 |
GO:0002940 |
tRNA N2-guanine methylation(GO:0002940) |
| 0.0 |
0.2 |
GO:0090527 |
actin filament reorganization(GO:0090527) |
| 0.0 |
0.1 |
GO:1904251 |
regulation of bile acid metabolic process(GO:1904251) |
| 0.0 |
0.1 |
GO:0048496 |
maintenance of organ identity(GO:0048496) |
| 0.0 |
0.1 |
GO:0001767 |
establishment of lymphocyte polarity(GO:0001767) establishment of T cell polarity(GO:0001768) |
| 0.0 |
0.2 |
GO:0036072 |
intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 |
0.1 |
GO:0015959 |
diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.0 |
0.1 |
GO:0007144 |
female meiosis I(GO:0007144) |
| 0.0 |
0.1 |
GO:0072385 |
minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.0 |
0.1 |
GO:0006011 |
UDP-glucose metabolic process(GO:0006011) |
| 0.0 |
0.3 |
GO:0018026 |
peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 |
0.3 |
GO:0006488 |
dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 |
0.1 |
GO:0090073 |
positive regulation of protein homodimerization activity(GO:0090073) positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.0 |
0.5 |
GO:0000920 |
cell separation after cytokinesis(GO:0000920) |
| 0.0 |
0.1 |
GO:0001955 |
blood vessel maturation(GO:0001955) |
| 0.0 |
0.1 |
GO:0072592 |
oxygen metabolic process(GO:0072592) |
| 0.0 |
0.1 |
GO:0032439 |
endosome localization(GO:0032439) |
| 0.0 |
0.1 |
GO:0071639 |
positive regulation of monocyte chemotactic protein-1 production(GO:0071639) |
| 0.0 |
0.1 |
GO:0045654 |
positive regulation of megakaryocyte differentiation(GO:0045654) positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.0 |
0.9 |
GO:1903846 |
positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 |
0.2 |
GO:0031441 |
negative regulation of mRNA 3'-end processing(GO:0031441) |
| 0.0 |
0.3 |
GO:0001731 |
formation of translation preinitiation complex(GO:0001731) |
| 0.0 |
0.3 |
GO:0030836 |
positive regulation of actin filament depolymerization(GO:0030836) |
| 0.0 |
0.2 |
GO:0006361 |
transcription initiation from RNA polymerase I promoter(GO:0006361) termination of RNA polymerase I transcription(GO:0006363) |
| 0.0 |
0.2 |
GO:1902847 |
macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of tau-protein kinase activity(GO:1902949) |
| 0.0 |
0.5 |
GO:0006103 |
2-oxoglutarate metabolic process(GO:0006103) |
| 0.0 |
0.3 |
GO:1902187 |
negative regulation of viral release from host cell(GO:1902187) |
| 0.0 |
0.2 |
GO:1903071 |
positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.0 |
0.3 |
GO:0051103 |
DNA ligation involved in DNA repair(GO:0051103) |
| 0.0 |
1.2 |
GO:0006446 |
regulation of translational initiation(GO:0006446) |
| 0.0 |
0.2 |
GO:0017196 |
N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 |
0.2 |
GO:0006384 |
transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 |
0.2 |
GO:0010561 |
negative regulation of glycoprotein biosynthetic process(GO:0010561) |
| 0.0 |
0.3 |
GO:0071786 |
endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 |
0.3 |
GO:1990173 |
protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) protein localization to nucleoplasm(GO:1990173) |
| 0.0 |
0.6 |
GO:1901998 |
toxin transport(GO:1901998) |
| 0.0 |
0.2 |
GO:0030206 |
chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 |
0.3 |
GO:0050932 |
regulation of melanocyte differentiation(GO:0045634) regulation of pigment cell differentiation(GO:0050932) |
| 0.0 |
0.3 |
GO:0048619 |
embryonic hindgut morphogenesis(GO:0048619) |
| 0.0 |
0.3 |
GO:0006622 |
protein targeting to lysosome(GO:0006622) |
| 0.0 |
0.1 |
GO:0016557 |
peroxisome membrane biogenesis(GO:0016557) |
| 0.0 |
0.4 |
GO:0045672 |
positive regulation of osteoclast differentiation(GO:0045672) |
| 0.0 |
0.4 |
GO:0006101 |
tricarboxylic acid cycle(GO:0006099) citrate metabolic process(GO:0006101) tricarboxylic acid metabolic process(GO:0072350) |
| 0.0 |
0.1 |
GO:0002315 |
marginal zone B cell differentiation(GO:0002315) |
| 0.0 |
0.1 |
GO:0034421 |
post-translational protein acetylation(GO:0034421) |
| 0.0 |
0.1 |
GO:0034047 |
regulation of protein phosphatase type 2A activity(GO:0034047) |
| 0.0 |
0.1 |
GO:0019243 |
methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 |
0.5 |
GO:0014823 |
response to activity(GO:0014823) |
| 0.0 |
0.5 |
GO:0044550 |
secondary metabolite biosynthetic process(GO:0044550) |
| 0.0 |
0.1 |
GO:0034975 |
protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 |
3.1 |
GO:0098780 |
mitophagy in response to mitochondrial depolarization(GO:0098779) response to mitochondrial depolarisation(GO:0098780) |
| 0.0 |
0.3 |
GO:0035721 |
intraciliary retrograde transport(GO:0035721) |
| 0.0 |
0.5 |
GO:2000785 |
regulation of autophagosome assembly(GO:2000785) |
| 0.0 |
0.1 |
GO:0019441 |
tryptophan catabolic process(GO:0006569) tryptophan catabolic process to kynurenine(GO:0019441) indole-containing compound catabolic process(GO:0042436) indolalkylamine catabolic process(GO:0046218) kynurenine metabolic process(GO:0070189) |
| 0.0 |
0.2 |
GO:0032780 |
negative regulation of ATPase activity(GO:0032780) |
| 0.0 |
0.3 |
GO:0045187 |
regulation of circadian sleep/wake cycle(GO:0042749) regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.0 |
0.1 |
GO:0006434 |
seryl-tRNA aminoacylation(GO:0006434) |
| 0.0 |
0.2 |
GO:0043950 |
positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.0 |
0.1 |
GO:0006436 |
tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.0 |
0.0 |
GO:0032286 |
central nervous system myelin maintenance(GO:0032286) |
| 0.0 |
0.3 |
GO:0048484 |
enteric nervous system development(GO:0048484) |
| 0.0 |
0.0 |
GO:1903748 |
negative regulation of establishment of protein localization to mitochondrion(GO:1903748) |
| 0.0 |
0.2 |
GO:0021775 |
smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.0 |
0.0 |
GO:0032510 |
endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.0 |
0.3 |
GO:0050832 |
defense response to fungus(GO:0050832) |
| 0.0 |
0.1 |
GO:1902268 |
negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 |
0.2 |
GO:0006654 |
phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.0 |
0.5 |
GO:0097352 |
autophagosome maturation(GO:0097352) |
| 0.0 |
0.1 |
GO:2000768 |
glomerular parietal epithelial cell differentiation(GO:0072139) positive regulation of epithelial cell differentiation involved in kidney development(GO:2000698) positive regulation of nephron tubule epithelial cell differentiation(GO:2000768) |
| 0.0 |
0.2 |
GO:0016480 |
negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.0 |
0.2 |
GO:0006691 |
leukotriene metabolic process(GO:0006691) |
| 0.0 |
0.0 |
GO:0000720 |
pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.0 |
0.3 |
GO:2000310 |
regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.0 |
0.1 |
GO:0061462 |
protein localization to lysosome(GO:0061462) |
| 0.0 |
0.1 |
GO:0043124 |
negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 |
0.2 |
GO:0007035 |
vacuolar acidification(GO:0007035) |
| 0.0 |
0.1 |
GO:0046645 |
positive regulation of gamma-delta T cell differentiation(GO:0045588) positive regulation of gamma-delta T cell activation(GO:0046645) |
| 0.0 |
0.1 |
GO:0090383 |
phagosome acidification(GO:0090383) |
| 0.0 |
0.1 |
GO:0021506 |
neural fold formation(GO:0001842) anterior neuropore closure(GO:0021506) neuropore closure(GO:0021995) |
| 0.0 |
0.3 |
GO:0016242 |
negative regulation of macroautophagy(GO:0016242) |
| 0.0 |
0.2 |
GO:2000114 |
regulation of establishment of cell polarity(GO:2000114) |
| 0.0 |
0.4 |
GO:0045742 |
positive regulation of epidermal growth factor receptor signaling pathway(GO:0045742) |
| 0.0 |
0.1 |
GO:1903361 |
protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 |
0.4 |
GO:2000311 |
regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 |
0.1 |
GO:0008635 |
activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.0 |
0.2 |
GO:0018095 |
protein polyglutamylation(GO:0018095) |
| 0.0 |
0.2 |
GO:0060009 |
Sertoli cell development(GO:0060009) |
| 0.0 |
0.1 |
GO:0010991 |
regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 |
0.2 |
GO:0042026 |
protein refolding(GO:0042026) |
| 0.0 |
0.6 |
GO:0006904 |
vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 |
0.1 |
GO:0050925 |
negative regulation of negative chemotaxis(GO:0050925) |
| 0.0 |
0.5 |
GO:0033108 |
mitochondrial respiratory chain complex assembly(GO:0033108) |
| 0.0 |
0.3 |
GO:0030213 |
hyaluronan biosynthetic process(GO:0030213) |
| 0.0 |
0.2 |
GO:0016322 |
neuron remodeling(GO:0016322) |
| 0.0 |
0.3 |
GO:0043248 |
proteasome assembly(GO:0043248) |
| 0.0 |
0.3 |
GO:0090201 |
negative regulation of release of cytochrome c from mitochondria(GO:0090201) |
| 0.0 |
0.3 |
GO:0042347 |
negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.0 |
0.1 |
GO:0006699 |
bile acid biosynthetic process(GO:0006699) |
| 0.0 |
0.1 |
GO:2000124 |
regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.0 |
0.2 |
GO:0007168 |
receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 |
0.4 |
GO:0046069 |
cGMP catabolic process(GO:0046069) |
| 0.0 |
1.0 |
GO:0032543 |
mitochondrial translation(GO:0032543) |
| 0.0 |
0.1 |
GO:0031293 |
membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 |
0.1 |
GO:0006896 |
Golgi to vacuole transport(GO:0006896) |
| 0.0 |
0.8 |
GO:0051897 |
positive regulation of protein kinase B signaling(GO:0051897) |
| 0.0 |
0.1 |
GO:0009957 |
epidermal cell fate specification(GO:0009957) |
| 0.0 |
0.2 |
GO:0020027 |
hemoglobin metabolic process(GO:0020027) |
| 0.0 |
0.0 |
GO:0051299 |
centrosome separation(GO:0051299) |
| 0.0 |
0.1 |
GO:0045647 |
negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 |
0.0 |
GO:0035188 |
blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 |
0.1 |
GO:0043486 |
histone exchange(GO:0043486) |
| 0.0 |
0.2 |
GO:0035563 |
positive regulation of chromatin binding(GO:0035563) |
| 0.0 |
0.2 |
GO:0060539 |
diaphragm development(GO:0060539) |
| 0.0 |
0.1 |
GO:0031860 |
telomeric 3' overhang formation(GO:0031860) |
| 0.0 |
0.1 |
GO:0043985 |
histone H4-R3 methylation(GO:0043985) |
| 0.0 |
0.1 |
GO:0048069 |
eye pigmentation(GO:0048069) |
| 0.0 |
0.1 |
GO:0042264 |
regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031585) peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 |
0.1 |
GO:0005981 |
regulation of glycogen catabolic process(GO:0005981) |
| 0.0 |
0.1 |
GO:0009136 |
ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.0 |
0.1 |
GO:0006481 |
C-terminal protein methylation(GO:0006481) |
| 0.0 |
0.0 |
GO:2000508 |
regulation of dendritic cell chemotaxis(GO:2000508) positive regulation of dendritic cell chemotaxis(GO:2000510) |
| 0.0 |
0.2 |
GO:0006661 |
phosphatidylinositol biosynthetic process(GO:0006661) |
| 0.0 |
0.3 |
GO:0030325 |
adrenal gland development(GO:0030325) |
| 0.0 |
0.1 |
GO:0006086 |
acetyl-CoA biosynthetic process from pyruvate(GO:0006086) |
| 0.0 |
0.1 |
GO:0033603 |
positive regulation of dopamine secretion(GO:0033603) |
| 0.0 |
0.1 |
GO:0006390 |
transcription from mitochondrial promoter(GO:0006390) |
| 0.0 |
0.0 |
GO:0098869 |
cellular oxidant detoxification(GO:0098869) |
| 0.0 |
0.0 |
GO:0031914 |
negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 |
0.2 |
GO:0010863 |
positive regulation of phospholipase C activity(GO:0010863) |
| 0.0 |
0.1 |
GO:0070375 |
ERK5 cascade(GO:0070375) |
| 0.0 |
0.1 |
GO:0043461 |
proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.0 |
0.1 |
GO:0023035 |
CD40 signaling pathway(GO:0023035) |
| 0.0 |
0.0 |
GO:0021747 |
cochlear nucleus development(GO:0021747) |
| 0.0 |
0.0 |
GO:1902915 |
negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.0 |
0.3 |
GO:0000303 |
response to superoxide(GO:0000303) |
| 0.0 |
0.0 |
GO:0014719 |
skeletal muscle satellite cell activation(GO:0014719) |
| 0.0 |
0.1 |
GO:0051546 |
vitamin A metabolic process(GO:0006776) positive regulation of skeletal muscle tissue regeneration(GO:0043415) regulation of skeletal muscle tissue regeneration(GO:0043416) keratinocyte migration(GO:0051546) |
| 0.0 |
0.1 |
GO:0006004 |
fucose metabolic process(GO:0006004) |
| 0.0 |
0.1 |
GO:0010719 |
negative regulation of epithelial to mesenchymal transition(GO:0010719) |
| 0.0 |
0.1 |
GO:0015888 |
thiamine transport(GO:0015888) |
| 0.0 |
0.1 |
GO:0016339 |
calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 |
0.1 |
GO:0032485 |
regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 |
0.2 |
GO:0071257 |
cellular response to electrical stimulus(GO:0071257) |
| 0.0 |
0.1 |
GO:0009081 |
branched-chain amino acid metabolic process(GO:0009081) branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 |
0.3 |
GO:0035873 |
lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.0 |
0.5 |
GO:0002230 |
positive regulation of defense response to virus by host(GO:0002230) |
| 0.0 |
0.1 |
GO:0036151 |
phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.0 |
0.0 |
GO:0008626 |
granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.0 |
0.1 |
GO:1902231 |
positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 |
0.1 |
GO:1902683 |
regulation of receptor localization to synapse(GO:1902683) |
| 0.0 |
0.1 |
GO:0051386 |
regulation of neurotrophin TRK receptor signaling pathway(GO:0051386) |
| 0.0 |
0.4 |
GO:0007218 |
neuropeptide signaling pathway(GO:0007218) |
| 0.0 |
0.2 |
GO:0060324 |
face development(GO:0060324) |
| 0.0 |
0.1 |
GO:0031274 |
positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 |
0.5 |
GO:0030317 |
sperm motility(GO:0030317) |
| 0.0 |
0.5 |
GO:2000134 |
negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 |
0.1 |
GO:0070050 |
neuron cellular homeostasis(GO:0070050) |
| 0.0 |
0.3 |
GO:0035335 |
peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 |
0.7 |
GO:0006406 |
mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 |
0.1 |
GO:1902236 |
negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.0 |
0.0 |
GO:0008612 |
peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.0 |
0.2 |
GO:0044342 |
type B pancreatic cell proliferation(GO:0044342) |
| 0.0 |
1.4 |
GO:0007030 |
Golgi organization(GO:0007030) |
| 0.0 |
0.1 |
GO:0006465 |
signal peptide processing(GO:0006465) |
| 0.0 |
0.3 |
GO:0007205 |
protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 |
0.0 |
GO:0042271 |
susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 |
0.1 |
GO:0043277 |
apoptotic cell clearance(GO:0043277) |
| 0.0 |
0.1 |
GO:0097150 |
neuronal stem cell population maintenance(GO:0097150) |
| 0.0 |
0.0 |
GO:1904154 |
positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.0 |
0.0 |
GO:0061086 |
negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 |
0.1 |
GO:0031100 |
organ regeneration(GO:0031100) |
| 0.0 |
0.2 |
GO:0032781 |
positive regulation of ATPase activity(GO:0032781) |
| 0.0 |
0.0 |
GO:0046084 |
'de novo' IMP biosynthetic process(GO:0006189) adenine metabolic process(GO:0046083) adenine biosynthetic process(GO:0046084) |
| 0.0 |
0.3 |
GO:0021904 |
dorsal/ventral neural tube patterning(GO:0021904) |
| 0.0 |
0.0 |
GO:0048311 |
mitochondrion distribution(GO:0048311) |
| 0.0 |
0.2 |
GO:0060325 |
face morphogenesis(GO:0060325) |
| 0.0 |
0.7 |
GO:0032526 |
response to retinoic acid(GO:0032526) |
| 0.0 |
0.1 |
GO:0035970 |
peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 |
0.1 |
GO:0098700 |
aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.0 |
0.2 |
GO:0006895 |
Golgi to endosome transport(GO:0006895) |
| 0.0 |
0.8 |
GO:0006334 |
nucleosome assembly(GO:0006334) |
| 0.0 |
0.2 |
GO:0010388 |
cullin deneddylation(GO:0010388) |
| 0.0 |
0.6 |
GO:0006888 |
ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 |
0.1 |
GO:0034983 |
peptidyl-lysine deacetylation(GO:0034983) |
| 0.0 |
0.3 |
GO:0060425 |
lung morphogenesis(GO:0060425) |
| 0.0 |
0.1 |
GO:0090344 |
negative regulation of cell aging(GO:0090344) |
| 0.0 |
0.1 |
GO:0045924 |
regulation of female receptivity(GO:0045924) |
| 0.0 |
0.2 |
GO:0008053 |
mitochondrial fusion(GO:0008053) |
| 0.0 |
0.3 |
GO:0010862 |
positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 |
0.0 |
GO:2000491 |
positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.0 |
0.3 |
GO:0031116 |
positive regulation of microtubule polymerization(GO:0031116) |
| 0.0 |
0.1 |
GO:0006878 |
cellular copper ion homeostasis(GO:0006878) |
| 0.0 |
0.2 |
GO:0033687 |
osteoblast proliferation(GO:0033687) |
| 0.0 |
0.1 |
GO:0090141 |
positive regulation of mitochondrial fission(GO:0090141) |
| 0.0 |
0.1 |
GO:0045002 |
double-strand break repair via single-strand annealing(GO:0045002) |
| 0.0 |
0.0 |
GO:0007021 |
tubulin complex assembly(GO:0007021) |
| 0.0 |
0.3 |
GO:0030901 |
midbrain development(GO:0030901) |
| 0.0 |
0.1 |
GO:0030049 |
muscle filament sliding(GO:0030049) |
| 0.0 |
0.4 |
GO:0001736 |
establishment of planar polarity(GO:0001736) establishment of tissue polarity(GO:0007164) |
| 0.0 |
0.1 |
GO:0009186 |
deoxyribonucleoside diphosphate metabolic process(GO:0009186) |
| 0.0 |
0.0 |
GO:0046381 |
CMP-N-acetylneuraminate metabolic process(GO:0046381) |
| 0.0 |
0.1 |
GO:0000103 |
sulfate assimilation(GO:0000103) |
| 0.0 |
0.1 |
GO:0018002 |
N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.0 |
0.1 |
GO:0060670 |
branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.0 |
0.0 |
GO:0048537 |
mucosal-associated lymphoid tissue development(GO:0048537) Peyer's patch development(GO:0048541) |
| 0.0 |
0.2 |
GO:0043388 |
positive regulation of DNA binding(GO:0043388) |
| 0.0 |
0.3 |
GO:0016578 |
histone deubiquitination(GO:0016578) |
| 0.0 |
0.1 |
GO:0070940 |
dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 |
0.1 |
GO:0045777 |
positive regulation of blood pressure(GO:0045777) |
| 0.0 |
0.0 |
GO:0035372 |
protein localization to microtubule(GO:0035372) |
| 0.0 |
0.3 |
GO:0006414 |
translational elongation(GO:0006414) |
| 0.0 |
0.0 |
GO:1990414 |
replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.0 |
0.0 |
GO:0002408 |
myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.0 |
0.1 |
GO:0010867 |
positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.0 |
0.1 |
GO:0050686 |
negative regulation of mRNA processing(GO:0050686) |
| 0.0 |
0.1 |
GO:0009052 |
pentose-phosphate shunt, non-oxidative branch(GO:0009052) |