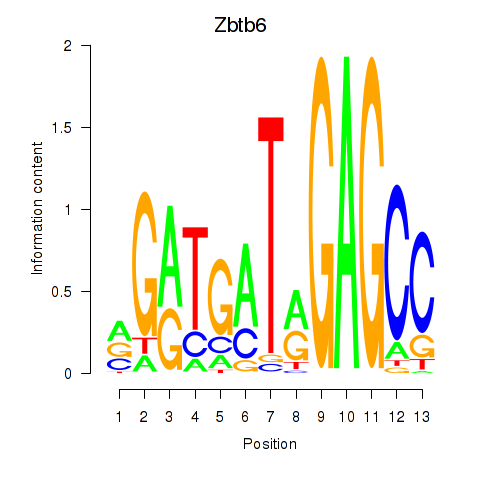
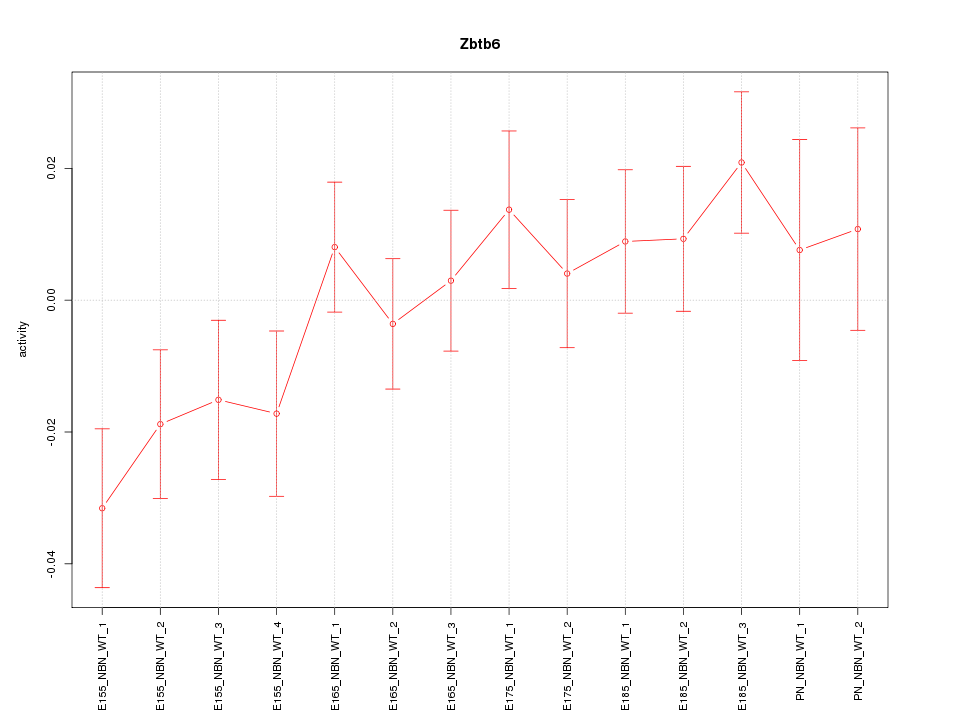
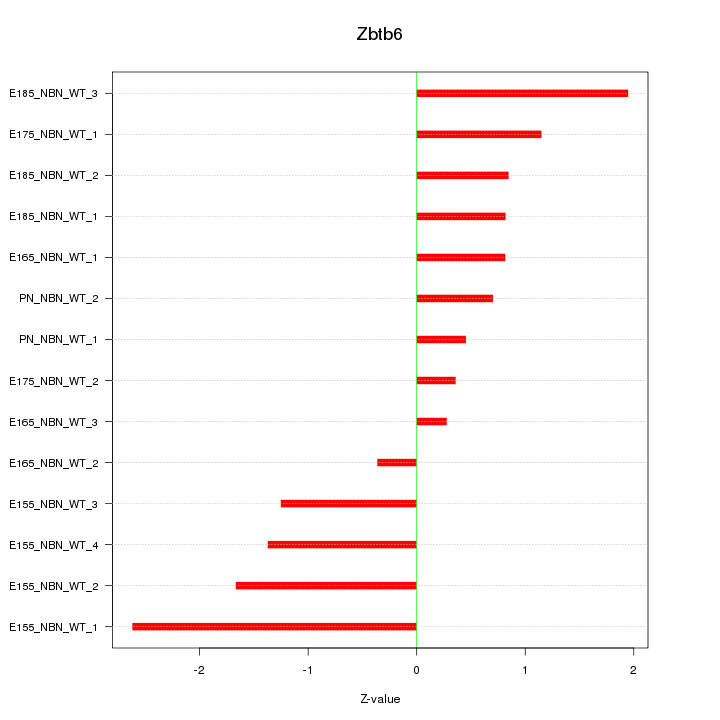
| 1.2 |
9.4 |
GO:1904714 |
regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.9 |
2.7 |
GO:0050929 |
corticospinal neuron axon guidance through spinal cord(GO:0021972) positive regulation of negative chemotaxis(GO:0050924) induction of negative chemotaxis(GO:0050929) negative regulation of mononuclear cell migration(GO:0071676) negative regulation of retinal ganglion cell axon guidance(GO:0090260) |
| 0.7 |
2.2 |
GO:0014063 |
negative regulation of serotonin secretion(GO:0014063) |
| 0.7 |
2.1 |
GO:0021837 |
motogenic signaling involved in postnatal olfactory bulb interneuron migration(GO:0021837) positive regulation of mitotic cell cycle DNA replication(GO:1903465) |
| 0.5 |
1.5 |
GO:2000224 |
testosterone biosynthetic process(GO:0061370) regulation of testosterone biosynthetic process(GO:2000224) |
| 0.5 |
1.5 |
GO:0061537 |
glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.4 |
1.6 |
GO:2000313 |
fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 0.4 |
1.1 |
GO:0030070 |
insulin processing(GO:0030070) |
| 0.4 |
1.9 |
GO:0032346 |
positive regulation of aldosterone metabolic process(GO:0032346) positive regulation of aldosterone biosynthetic process(GO:0032349) regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.4 |
7.4 |
GO:0015812 |
gamma-aminobutyric acid transport(GO:0015812) |
| 0.4 |
1.1 |
GO:2000564 |
CD8-positive, alpha-beta T cell proliferation(GO:0035740) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) positive regulation of CD8-positive, alpha-beta T cell activation(GO:2001187) |
| 0.4 |
1.1 |
GO:1904706 |
response to cisplatin(GO:0072718) negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.3 |
1.0 |
GO:0061743 |
motor learning(GO:0061743) |
| 0.3 |
2.1 |
GO:0036006 |
response to macrophage colony-stimulating factor(GO:0036005) cellular response to macrophage colony-stimulating factor stimulus(GO:0036006) |
| 0.3 |
1.2 |
GO:0006547 |
histidine metabolic process(GO:0006547) imidazole-containing compound metabolic process(GO:0052803) |
| 0.3 |
0.8 |
GO:0030321 |
transepithelial chloride transport(GO:0030321) transepithelial ammonium transport(GO:0070634) |
| 0.3 |
0.8 |
GO:1900158 |
negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.3 |
1.0 |
GO:0072137 |
condensed mesenchymal cell proliferation(GO:0072137) |
| 0.2 |
0.7 |
GO:0071420 |
cellular response to histamine(GO:0071420) |
| 0.2 |
1.5 |
GO:0033353 |
S-adenosylmethionine cycle(GO:0033353) |
| 0.2 |
1.3 |
GO:0016338 |
calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.2 |
1.1 |
GO:0050908 |
detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.2 |
0.5 |
GO:0003278 |
apoptotic process involved in heart morphogenesis(GO:0003278) |
| 0.2 |
1.7 |
GO:2001214 |
positive regulation of vasculogenesis(GO:2001214) |
| 0.2 |
1.0 |
GO:0032483 |
regulation of Rab protein signal transduction(GO:0032483) |
| 0.2 |
0.7 |
GO:0001907 |
killing by symbiont of host cells(GO:0001907) induction of programmed cell death(GO:0012502) disruption by symbiont of host cell(GO:0044004) positive regulation of apoptotic process in other organism(GO:0044533) positive regulation by symbiont of host programmed cell death(GO:0052042) positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052330) positive regulation by organism of apoptotic process in other organism involved in symbiotic interaction(GO:0052501) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.1 |
0.4 |
GO:0006601 |
creatine biosynthetic process(GO:0006601) |
| 0.1 |
1.3 |
GO:0097151 |
spontaneous neurotransmitter secretion(GO:0061669) positive regulation of inhibitory postsynaptic potential(GO:0097151) spontaneous synaptic transmission(GO:0098814) |
| 0.1 |
3.8 |
GO:1902476 |
chloride transmembrane transport(GO:1902476) |
| 0.1 |
0.3 |
GO:0036500 |
ATF6-mediated unfolded protein response(GO:0036500) |
| 0.1 |
1.3 |
GO:0042759 |
long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.1 |
0.6 |
GO:0031119 |
tRNA pseudouridine synthesis(GO:0031119) |
| 0.1 |
0.2 |
GO:0060743 |
epithelial cell maturation involved in prostate gland development(GO:0060743) alveolar secondary septum development(GO:0061144) |
| 0.1 |
1.3 |
GO:0044243 |
collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.1 |
0.6 |
GO:0009404 |
toxin metabolic process(GO:0009404) |
| 0.1 |
0.2 |
GO:0019478 |
D-amino acid catabolic process(GO:0019478) |
| 0.1 |
0.4 |
GO:2000348 |
regulation of CD40 signaling pathway(GO:2000348) |
| 0.1 |
0.2 |
GO:0070172 |
oculomotor nerve development(GO:0021557) positive regulation of tooth mineralization(GO:0070172) |
| 0.1 |
0.5 |
GO:0032020 |
ISG15-protein conjugation(GO:0032020) |
| 0.0 |
0.2 |
GO:1901678 |
iron coordination entity transport(GO:1901678) |
| 0.0 |
0.4 |
GO:0070475 |
rRNA base methylation(GO:0070475) |
| 0.0 |
0.6 |
GO:0060081 |
membrane hyperpolarization(GO:0060081) |
| 0.0 |
0.6 |
GO:0016486 |
peptide hormone processing(GO:0016486) |
| 0.0 |
0.3 |
GO:0038026 |
reelin-mediated signaling pathway(GO:0038026) |
| 0.0 |
0.3 |
GO:0042118 |
blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) endothelial cell activation(GO:0042118) |
| 0.0 |
0.1 |
GO:0046340 |
diacylglycerol catabolic process(GO:0046340) |
| 0.0 |
1.1 |
GO:0045601 |
regulation of endothelial cell differentiation(GO:0045601) |
| 0.0 |
1.0 |
GO:0030318 |
melanocyte differentiation(GO:0030318) |
| 0.0 |
0.2 |
GO:0060272 |
embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.0 |
0.4 |
GO:0000012 |
single strand break repair(GO:0000012) |
| 0.0 |
0.2 |
GO:0005981 |
regulation of glycogen catabolic process(GO:0005981) |
| 0.0 |
0.1 |
GO:0071550 |
death-inducing signaling complex assembly(GO:0071550) |
| 0.0 |
0.9 |
GO:0030199 |
collagen fibril organization(GO:0030199) |
| 0.0 |
0.2 |
GO:0046543 |
development of secondary female sexual characteristics(GO:0046543) |
| 0.0 |
0.4 |
GO:0071577 |
zinc II ion transmembrane transport(GO:0071577) |
| 0.0 |
0.4 |
GO:0001553 |
luteinization(GO:0001553) |
| 0.0 |
0.2 |
GO:0071318 |
cellular response to ATP(GO:0071318) |
| 0.0 |
0.1 |
GO:1903361 |
protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 |
1.8 |
GO:0043488 |
regulation of mRNA stability(GO:0043488) |
| 0.0 |
0.1 |
GO:2000491 |
positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.0 |
0.7 |
GO:0002474 |
antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 |
0.4 |
GO:0046677 |
response to antibiotic(GO:0046677) |
| 0.0 |
0.4 |
GO:0040034 |
regulation of development, heterochronic(GO:0040034) |
| 0.0 |
0.4 |
GO:0071158 |
positive regulation of cell cycle arrest(GO:0071158) |
| 0.0 |
0.1 |
GO:0021957 |
corticospinal tract morphogenesis(GO:0021957) |
| 0.0 |
0.4 |
GO:0043949 |
regulation of cAMP-mediated signaling(GO:0043949) |
| 0.0 |
0.4 |
GO:0034724 |
DNA replication-independent nucleosome assembly(GO:0006336) DNA replication-independent nucleosome organization(GO:0034724) |
| 0.0 |
0.4 |
GO:0032728 |
positive regulation of interferon-beta production(GO:0032728) |
| 0.0 |
0.1 |
GO:2000465 |
regulation of glycogen (starch) synthase activity(GO:2000465) |
| 0.0 |
0.3 |
GO:0055012 |
ventricular cardiac muscle cell differentiation(GO:0055012) |