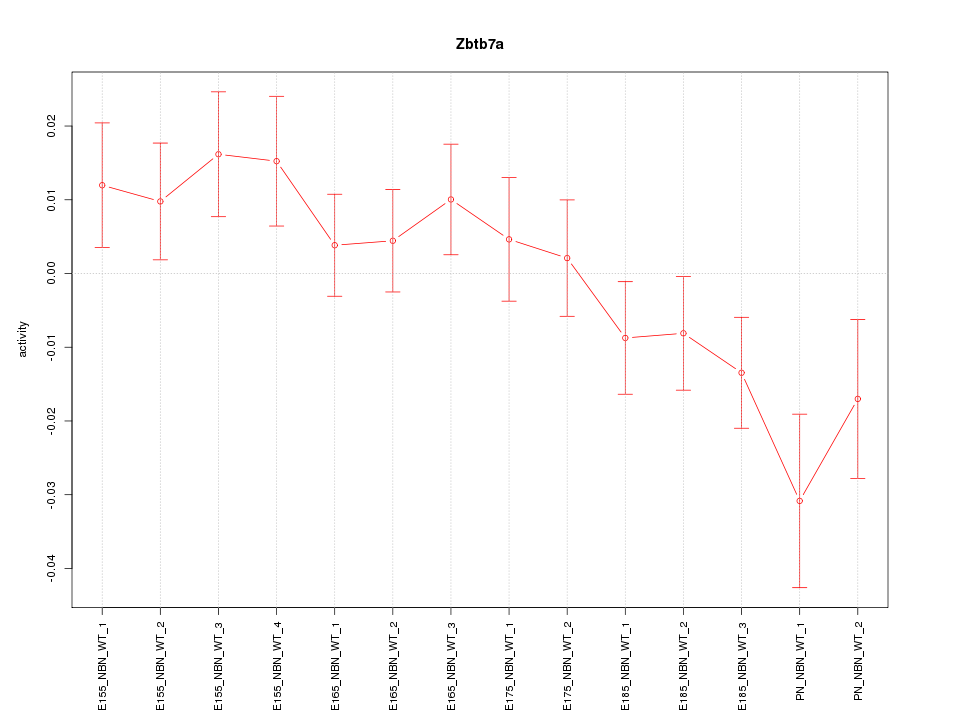
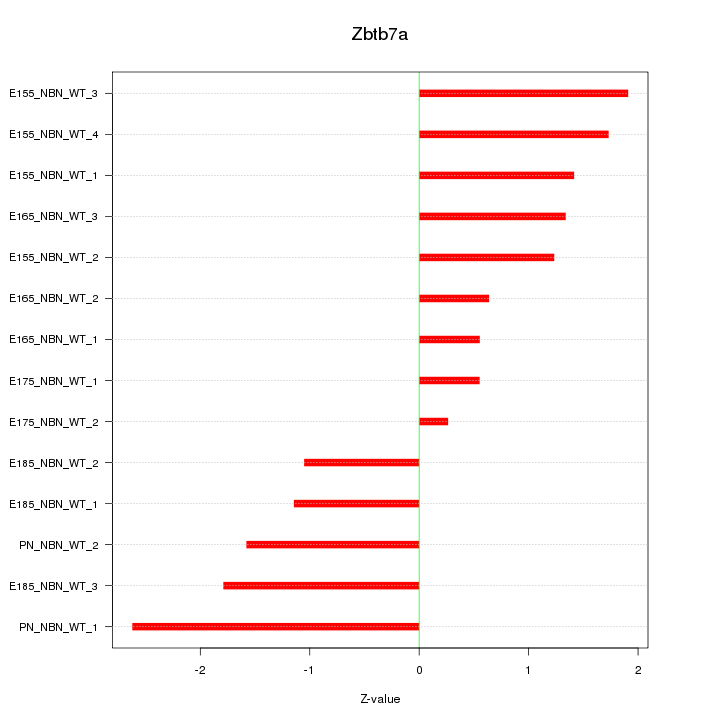
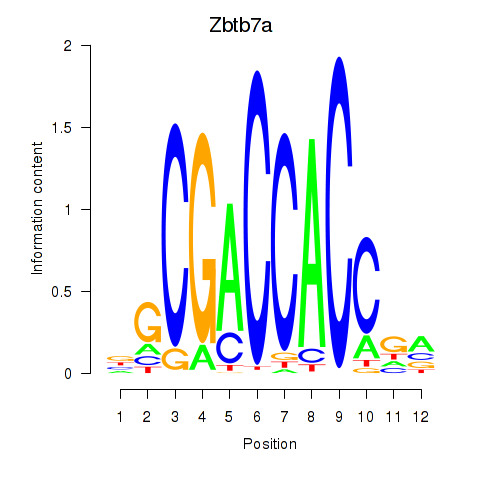
Motif ID: Zbtb7a
Z-value: 1.415
Transcription factors associated with Zbtb7a:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Zbtb7a | ENSMUSG00000035011.9 | Zbtb7a |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zbtb7a | mm10_v2_chr10_+_81136223_81136271 | -0.29 | 3.1e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.7 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 1.1 | 4.5 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.9 | 2.8 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 0.9 | 3.5 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.8 | 3.2 | GO:0060915 | fibroblast growth factor receptor signaling pathway involved in mammary gland specification(GO:0060595) mammary gland bud formation(GO:0060615) branch elongation involved in salivary gland morphogenesis(GO:0060667) mesenchymal cell differentiation involved in lung development(GO:0060915) |
| 0.8 | 3.9 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.7 | 2.8 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.6 | 1.9 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 0.6 | 2.2 | GO:0061526 | acetylcholine secretion, neurotransmission(GO:0014055) regulation of acetylcholine secretion, neurotransmission(GO:0014056) acetylcholine secretion(GO:0061526) |
| 0.6 | 2.2 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.5 | 3.0 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.5 | 2.5 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) |
| 0.5 | 1.8 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.4 | 1.7 | GO:0060912 | cardiac cell fate specification(GO:0060912) |
| 0.4 | 2.1 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.4 | 2.4 | GO:1901524 | regulation of macromitophagy(GO:1901524) negative regulation of macromitophagy(GO:1901525) |
| 0.4 | 1.2 | GO:1904742 | protein poly-ADP-ribosylation(GO:0070212) regulation of telomeric DNA binding(GO:1904742) |
| 0.4 | 3.5 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.4 | 1.1 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.4 | 1.1 | GO:0003289 | atrial septum primum morphogenesis(GO:0003289) |
| 0.4 | 2.3 | GO:0010917 | negative regulation of mitochondrial membrane potential(GO:0010917) |
| 0.4 | 1.8 | GO:0048631 | regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.3 | 1.0 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.3 | 1.0 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 0.3 | 4.7 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.3 | 1.5 | GO:0034436 | glycoprotein transport(GO:0034436) |
| 0.3 | 2.6 | GO:0002536 | respiratory burst involved in inflammatory response(GO:0002536) |
| 0.3 | 0.9 | GO:0060598 | dichotomous subdivision of terminal units involved in mammary gland duct morphogenesis(GO:0060598) |
| 0.3 | 1.0 | GO:0045343 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) positive regulation of MHC class I biosynthetic process(GO:0045345) |
| 0.3 | 1.0 | GO:0021564 | glossopharyngeal nerve development(GO:0021563) vagus nerve development(GO:0021564) |
| 0.2 | 1.7 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.2 | 0.7 | GO:2000744 | anterior head development(GO:0097065) regulation of anterior head development(GO:2000742) positive regulation of anterior head development(GO:2000744) |
| 0.2 | 0.9 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.2 | 0.7 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.2 | 0.7 | GO:0046381 | CMP-N-acetylneuraminate metabolic process(GO:0046381) |
| 0.2 | 1.8 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.2 | 0.9 | GO:0003149 | membranous septum morphogenesis(GO:0003149) |
| 0.2 | 1.9 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.2 | 1.0 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.2 | 0.6 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.2 | 1.6 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.2 | 0.8 | GO:0006203 | dGTP catabolic process(GO:0006203) dATP catabolic process(GO:0046061) |
| 0.2 | 0.7 | GO:0003360 | brainstem development(GO:0003360) |
| 0.2 | 0.5 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylserine scrambling(GO:0061589) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.2 | 1.4 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.2 | 0.5 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.2 | 0.7 | GO:0046084 | adenine metabolic process(GO:0046083) adenine biosynthetic process(GO:0046084) |
| 0.2 | 0.8 | GO:0051012 | microtubule sliding(GO:0051012) negative regulation of nonmotile primary cilium assembly(GO:1902856) |
| 0.2 | 0.5 | GO:0003134 | BMP signaling pathway involved in heart induction(GO:0003130) endodermal-mesodermal cell signaling(GO:0003133) endodermal-mesodermal cell signaling involved in heart induction(GO:0003134) mesenchymal cell proliferation involved in ureteric bud development(GO:0072138) |
| 0.2 | 1.0 | GO:0051790 | short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.2 | 0.5 | GO:2000170 | positive regulation of peptidyl-cysteine S-nitrosylation(GO:2000170) |
| 0.2 | 1.2 | GO:1904217 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) regulation of serine C-palmitoyltransferase activity(GO:1904220) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.2 | 0.8 | GO:0060398 | regulation of growth hormone receptor signaling pathway(GO:0060398) |
| 0.2 | 0.5 | GO:0032650 | regulation of interleukin-1 alpha production(GO:0032650) positive regulation of interleukin-1 alpha production(GO:0032730) interleukin-1 alpha secretion(GO:0050703) |
| 0.2 | 2.4 | GO:0021756 | striatum development(GO:0021756) |
| 0.2 | 0.5 | GO:0032240 | negative regulation of nucleobase-containing compound transport(GO:0032240) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.2 | 0.5 | GO:0032058 | positive regulation of translational initiation in response to stress(GO:0032058) |
| 0.2 | 2.0 | GO:0090331 | negative regulation of platelet aggregation(GO:0090331) |
| 0.2 | 0.5 | GO:0014054 | positive regulation of gamma-aminobutyric acid secretion(GO:0014054) |
| 0.2 | 1.1 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.2 | 0.6 | GO:1904008 | response to monosodium glutamate(GO:1904008) cellular response to monosodium glutamate(GO:1904009) |
| 0.2 | 0.5 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.1 | 2.1 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.1 | 0.4 | GO:1900060 | negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.1 | 1.3 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.1 | 1.4 | GO:1901409 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.1 | 1.0 | GO:0046855 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) |
| 0.1 | 0.7 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.1 | 0.7 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.1 | 0.5 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.1 | 0.5 | GO:0019355 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.1 | 1.7 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.1 | 0.5 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.1 | 1.4 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.1 | 0.4 | GO:0048211 | Golgi vesicle docking(GO:0048211) |
| 0.1 | 0.4 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.1 | 0.4 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.1 | 0.8 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.1 | 0.7 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.1 | 1.5 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.1 | 1.8 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.1 | 0.7 | GO:0001302 | replicative cell aging(GO:0001302) |
| 0.1 | 1.1 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.1 | 0.4 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.1 | 6.7 | GO:0015807 | L-amino acid transport(GO:0015807) |
| 0.1 | 0.5 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.1 | 0.5 | GO:0045759 | negative regulation of action potential(GO:0045759) |
| 0.1 | 3.8 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.1 | 0.7 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.1 | 0.6 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 1.3 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.1 | 0.3 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.1 | 0.8 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.1 | 0.3 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.1 | 0.6 | GO:0061052 | negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 0.1 | 2.7 | GO:0042059 | negative regulation of epidermal growth factor receptor signaling pathway(GO:0042059) |
| 0.1 | 0.5 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 1.4 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.1 | 0.6 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.1 | 0.4 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.1 | 0.5 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 1.1 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.1 | 1.3 | GO:0061339 | establishment of monopolar cell polarity(GO:0061162) establishment or maintenance of monopolar cell polarity(GO:0061339) |
| 0.1 | 1.0 | GO:0070445 | oligodendrocyte progenitor proliferation(GO:0070444) regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.1 | 2.0 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 1.4 | GO:0021533 | cell differentiation in hindbrain(GO:0021533) |
| 0.1 | 1.8 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.1 | 2.3 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.1 | 0.7 | GO:0034312 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) sphingoid biosynthetic process(GO:0046520) |
| 0.1 | 0.3 | GO:0060527 | prostate glandular acinus morphogenesis(GO:0060526) prostate epithelial cord arborization involved in prostate glandular acinus morphogenesis(GO:0060527) |
| 0.1 | 0.1 | GO:0035984 | response to trichostatin A(GO:0035983) cellular response to trichostatin A(GO:0035984) |
| 0.1 | 0.8 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.1 | 0.3 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) negative regulation of neutrophil activation(GO:1902564) |
| 0.1 | 0.5 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.1 | 0.6 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.1 | 0.3 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.1 | 0.5 | GO:0010587 | miRNA catabolic process(GO:0010587) nuclear-transcribed mRNA catabolic process, exonucleolytic, 3'-5'(GO:0034427) |
| 0.1 | 5.1 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.1 | 0.2 | GO:0070970 | interleukin-2 secretion(GO:0070970) |
| 0.1 | 0.2 | GO:0032532 | regulation of microvillus length(GO:0032532) |
| 0.1 | 0.8 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.1 | 1.2 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.1 | 1.5 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.1 | 0.1 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.1 | 1.0 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.1 | 0.4 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.1 | 0.4 | GO:0015684 | ferrous iron transport(GO:0015684) |
| 0.1 | 4.3 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 0.7 | GO:0046835 | carbohydrate phosphorylation(GO:0046835) |
| 0.0 | 1.4 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.0 | 0.7 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 0.1 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.0 | 0.2 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 0.8 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.0 | 0.7 | GO:0060213 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.0 | 0.2 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.0 | 0.0 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.0 | 0.2 | GO:0060330 | regulation of response to interferon-gamma(GO:0060330) |
| 0.0 | 0.5 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 0.1 | GO:2000646 | positive regulation of receptor catabolic process(GO:2000646) |
| 0.0 | 0.4 | GO:0034122 | negative regulation of toll-like receptor signaling pathway(GO:0034122) |
| 0.0 | 0.6 | GO:0021957 | corticospinal tract morphogenesis(GO:0021957) |
| 0.0 | 0.7 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 1.4 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.3 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.0 | 0.3 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.0 | 0.1 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.0 | 0.9 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.0 | 0.1 | GO:0034382 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) |
| 0.0 | 0.7 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.0 | 0.5 | GO:0051352 | negative regulation of ligase activity(GO:0051352) negative regulation of ubiquitin-protein transferase activity(GO:0051444) |
| 0.0 | 0.6 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.2 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.1 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.0 | 0.1 | GO:0031126 | snoRNA 3'-end processing(GO:0031126) |
| 0.0 | 0.1 | GO:2000109 | macrophage apoptotic process(GO:0071888) scaRNA localization to Cajal body(GO:0090666) regulation of macrophage apoptotic process(GO:2000109) |
| 0.0 | 0.1 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.0 | 0.5 | GO:0033198 | response to ATP(GO:0033198) |
| 0.0 | 3.0 | GO:0043433 | negative regulation of sequence-specific DNA binding transcription factor activity(GO:0043433) |
| 0.0 | 0.6 | GO:0009409 | response to cold(GO:0009409) |
| 0.0 | 0.3 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) regulation of histone H3-K9 trimethylation(GO:1900112) negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 0.1 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.0 | GO:0042275 | error-free postreplication DNA repair(GO:0042275) |
| 0.0 | 0.4 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 0.1 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.0 | 0.2 | GO:0090244 | Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.0 | 0.2 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 1.3 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.0 | 0.1 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.0 | 0.2 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 1.9 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 0.3 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.0 | 0.1 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.1 | GO:0002553 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.0 | 0.3 | GO:0036010 | protein localization to endosome(GO:0036010) |
| 0.0 | 0.3 | GO:0006744 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.1 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.0 | 0.4 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.3 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.4 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.7 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 0.1 | GO:0097343 | ripoptosome assembly(GO:0097343) ripoptosome assembly involved in necroptotic process(GO:1901026) |
| 0.0 | 0.6 | GO:0048635 | negative regulation of muscle organ development(GO:0048635) |
| 0.0 | 0.2 | GO:0006337 | nucleosome disassembly(GO:0006337) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 0.7 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.1 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 2.4 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.0 | 0.2 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 | 0.1 | GO:0061684 | chaperone-mediated autophagy(GO:0061684) |
| 0.0 | 0.4 | GO:0044804 | nucleophagy(GO:0044804) |
| 0.0 | 0.4 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.4 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.2 | GO:0002183 | cytoplasmic translational initiation(GO:0002183) |
| 0.0 | 0.2 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.1 | GO:0035067 | negative regulation of histone acetylation(GO:0035067) |
| 0.0 | 0.5 | GO:0035019 | somatic stem cell population maintenance(GO:0035019) |
| 0.0 | 0.1 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.0 | 0.2 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.4 | GO:0031214 | biomineral tissue development(GO:0031214) |
| 0.0 | 0.4 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.7 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 0.1 | GO:0018065 | protein-cofactor linkage(GO:0018065) |
| 0.0 | 0.8 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.6 | GO:0070373 | negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.0 | 0.3 | GO:0043330 | response to exogenous dsRNA(GO:0043330) |
| 0.0 | 0.2 | GO:1902042 | negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.0 | 0.5 | GO:0035418 | protein localization to synapse(GO:0035418) |
| 0.0 | 0.6 | GO:0051865 | protein autoubiquitination(GO:0051865) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.7 | GO:0090537 | CERF complex(GO:0090537) |
| 0.4 | 1.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.4 | 3.6 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.3 | 2.1 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.3 | 5.7 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.2 | 0.7 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
| 0.2 | 2.3 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.2 | 3.0 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.2 | 0.5 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.1 | 2.8 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 0.4 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.1 | 0.8 | GO:0000938 | GARP complex(GO:0000938) |
| 0.1 | 1.5 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 0.4 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.1 | 0.5 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.1 | 0.4 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 1.9 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.1 | 0.7 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.1 | 0.7 | GO:0000836 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.1 | 0.4 | GO:0005712 | chiasma(GO:0005712) |
| 0.1 | 0.5 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.1 | 1.3 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 0.8 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.1 | 0.6 | GO:0005638 | lamin filament(GO:0005638) |
| 0.1 | 1.4 | GO:0034719 | SMN-Sm protein complex(GO:0034719) |
| 0.1 | 0.6 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.1 | 0.6 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 0.6 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 0.7 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.1 | 0.5 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 2.6 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 0.8 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 0.3 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.1 | 3.7 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 6.4 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 1.3 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 0.4 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.1 | 1.2 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 0.5 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 1.1 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.5 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.4 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.0 | 0.2 | GO:0043259 | laminin-1 complex(GO:0005606) laminin-10 complex(GO:0043259) |
| 0.0 | 1.0 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.6 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.5 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 3.5 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.3 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.0 | 0.1 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 0.5 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.4 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.8 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.3 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.7 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.2 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.1 | GO:0031088 | platelet dense granule membrane(GO:0031088) lysosomal lumen(GO:0043202) |
| 0.0 | 0.1 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.0 | 0.3 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 2.4 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 0.1 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.6 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.4 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.2 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.3 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.6 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.2 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.3 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.4 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 1.0 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 1.3 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 2.5 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 0.5 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.1 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 1.1 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 1.1 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.1 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.4 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.7 | GO:0031228 | intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.4 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.7 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.4 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 1.2 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 1.0 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.1 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 0.1 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.0 | 0.6 | GO:0000791 | euchromatin(GO:0000791) |
| 0.0 | 2.1 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.1 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.3 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.4 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.9 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.7 | 2.2 | GO:0001565 | phorbol ester receptor activity(GO:0001565) non-kinase phorbol ester receptor activity(GO:0001566) |
| 0.6 | 3.5 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) lysophosphatidic acid receptor activity(GO:0070915) |
| 0.6 | 2.2 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) serotonin receptor activity(GO:0099589) |
| 0.5 | 1.5 | GO:0034189 | very-low-density lipoprotein particle binding(GO:0034189) glycoprotein transporter activity(GO:0034437) |
| 0.5 | 1.9 | GO:1902379 | chemoattractant activity involved in axon guidance(GO:1902379) |
| 0.4 | 3.0 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.4 | 1.5 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.3 | 1.0 | GO:0004315 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) |
| 0.2 | 1.0 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.2 | 1.0 | GO:0004441 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) |
| 0.2 | 0.7 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.2 | 1.9 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) |
| 0.2 | 1.6 | GO:0015198 | oligopeptide transporter activity(GO:0015198) |
| 0.2 | 2.6 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.2 | 0.7 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.2 | 0.4 | GO:0004676 | 3-phosphoinositide-dependent protein kinase activity(GO:0004676) |
| 0.2 | 1.0 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.2 | 5.6 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.2 | 0.6 | GO:0047256 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.2 | 8.1 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.2 | 3.0 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.2 | 2.5 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.2 | 1.3 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.2 | 2.8 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.2 | 0.5 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.1 | 2.2 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 4.5 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 3.3 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 0.7 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.1 | 0.5 | GO:0004515 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.1 | 0.8 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.1 | 0.7 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 0.7 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.1 | 0.7 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.1 | 1.6 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.1 | 0.6 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.1 | 5.2 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.1 | 1.3 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.1 | 1.9 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.1 | 0.5 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 0.8 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 1.0 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 1.1 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 1.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 0.5 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 0.3 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.1 | 0.7 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) |
| 0.1 | 0.3 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) dinucleotide insertion or deletion binding(GO:0032139) single guanine insertion binding(GO:0032142) |
| 0.1 | 0.6 | GO:0098518 | polynucleotide phosphatase activity(GO:0098518) |
| 0.1 | 0.6 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.1 | 0.3 | GO:0035473 | lipase binding(GO:0035473) |
| 0.1 | 0.5 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.1 | 1.0 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.1 | 1.5 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 0.5 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 0.3 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 1.1 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.1 | 0.4 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.1 | 1.8 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.1 | 0.5 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.1 | 0.4 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.1 | 1.4 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 0.9 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 0.6 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 0.5 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.1 | 2.0 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 0.8 | GO:0032554 | purine deoxyribonucleotide binding(GO:0032554) |
| 0.1 | 1.0 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.1 | 0.7 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.1 | 0.6 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.1 | 0.4 | GO:0015087 | cobalt ion transmembrane transporter activity(GO:0015087) |
| 0.1 | 0.6 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 1.4 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 0.2 | GO:0018121 | imidazoleglycerol-phosphate synthase activity(GO:0000107) NAD(P)-cysteine ADP-ribosyltransferase activity(GO:0018071) NAD(P)-asparagine ADP-ribosyltransferase activity(GO:0018121) NAD(P)-serine ADP-ribosyltransferase activity(GO:0018127) 7-cyano-7-deazaguanine tRNA-ribosyltransferase activity(GO:0043867) purine deoxyribosyltransferase activity(GO:0044102) |
| 0.0 | 0.5 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.1 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.2 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.7 | GO:0052813 | phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.7 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.0 | 0.3 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.3 | GO:0051718 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates(GO:0051718) |
| 0.0 | 0.9 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.3 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.0 | 3.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.2 | GO:0035325 | Toll-like receptor binding(GO:0035325) |
| 0.0 | 0.7 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.5 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.3 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.2 | GO:0052833 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.2 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.0 | 1.2 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 2.1 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 1.5 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.6 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 1.2 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.6 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 4.0 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 1.1 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.3 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.5 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.2 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.6 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.5 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 0.1 | GO:0030362 | protein phosphatase type 4 regulator activity(GO:0030362) |
| 0.0 | 0.1 | GO:0016361 | activin receptor activity, type I(GO:0016361) activin-activated receptor activity(GO:0017002) |
| 0.0 | 0.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.2 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.1 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 0.0 | 0.5 | GO:0018602 | sulfonate dioxygenase activity(GO:0000907) 2,4-dichlorophenoxyacetate alpha-ketoglutarate dioxygenase activity(GO:0018602) hypophosphite dioxygenase activity(GO:0034792) gibberellin 2-beta-dioxygenase activity(GO:0045543) C-19 gibberellin 2-beta-dioxygenase activity(GO:0052634) C-20 gibberellin 2-beta-dioxygenase activity(GO:0052635) |
| 0.0 | 1.7 | GO:0043851 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) rRNA (uridine-2'-O-)-methyltransferase activity(GO:0008650) rRNA (adenine-N6-)-methyltransferase activity(GO:0008988) DNA-methyltransferase activity(GO:0009008) rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) selenocysteine methyltransferase activity(GO:0016205) rRNA (adenine) methyltransferase activity(GO:0016433) rRNA (cytosine) methyltransferase activity(GO:0016434) rRNA (guanine) methyltransferase activity(GO:0016435) 1-phenanthrol methyltransferase activity(GO:0018707) protein-arginine N5-methyltransferase activity(GO:0019702) dimethylarsinite methyltransferase activity(GO:0034541) 4,5-dihydroxybenzo(a)pyrene methyltransferase activity(GO:0034807) 1-hydroxypyrene methyltransferase activity(GO:0034931) 1-hydroxy-6-methoxypyrene methyltransferase activity(GO:0034933) demethylmenaquinone methyltransferase activity(GO:0043770) cobalt-precorrin-6B C5-methyltransferase activity(GO:0043776) cobalt-precorrin-7 C15-methyltransferase activity(GO:0043777) cobalt-precorrin-5B C1-methyltransferase activity(GO:0043780) cobalt-precorrin-3 C17-methyltransferase activity(GO:0043782) dimethylamine methyltransferase activity(GO:0043791) hydroxyneurosporene-O-methyltransferase activity(GO:0043803) tRNA (adenine-57, 58-N(1)-) methyltransferase activity(GO:0043827) methylamine-specific methylcobalamin:coenzyme M methyltransferase activity(GO:0043833) trimethylamine methyltransferase activity(GO:0043834) methanol-specific methylcobalamin:coenzyme M methyltransferase activity(GO:0043851) monomethylamine methyltransferase activity(GO:0043852) P-methyltransferase activity(GO:0051994) Se-methyltransferase activity(GO:0051995) 2-phytyl-1,4-naphthoquinone methyltransferase activity(GO:0052624) tRNA (uracil-2'-O-)-methyltransferase activity(GO:0052665) tRNA (cytosine-2'-O-)-methyltransferase activity(GO:0052666) phosphomethylethanolamine N-methyltransferase activity(GO:0052667) tRNA (cytosine-3-)-methyltransferase activity(GO:0052735) rRNA (cytosine-2'-O-)-methyltransferase activity(GO:0070677) rRNA (cytosine-N4-)-methyltransferase activity(GO:0071424) trihydroxyferuloyl spermidine O-methyltransferase activity(GO:0080012) |
| 0.0 | 0.1 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.8 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.2 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.6 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 1.7 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 2.2 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.7 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.4 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) |
| 0.0 | 2.4 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.0 | 0.1 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.4 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.2 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.3 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.1 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 1.8 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.0 | 1.2 | GO:0004721 | phosphoprotein phosphatase activity(GO:0004721) |
| 0.0 | 5.0 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 0.1 | GO:0004661 | protein geranylgeranyltransferase activity(GO:0004661) |
| 0.0 | 0.2 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.4 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |