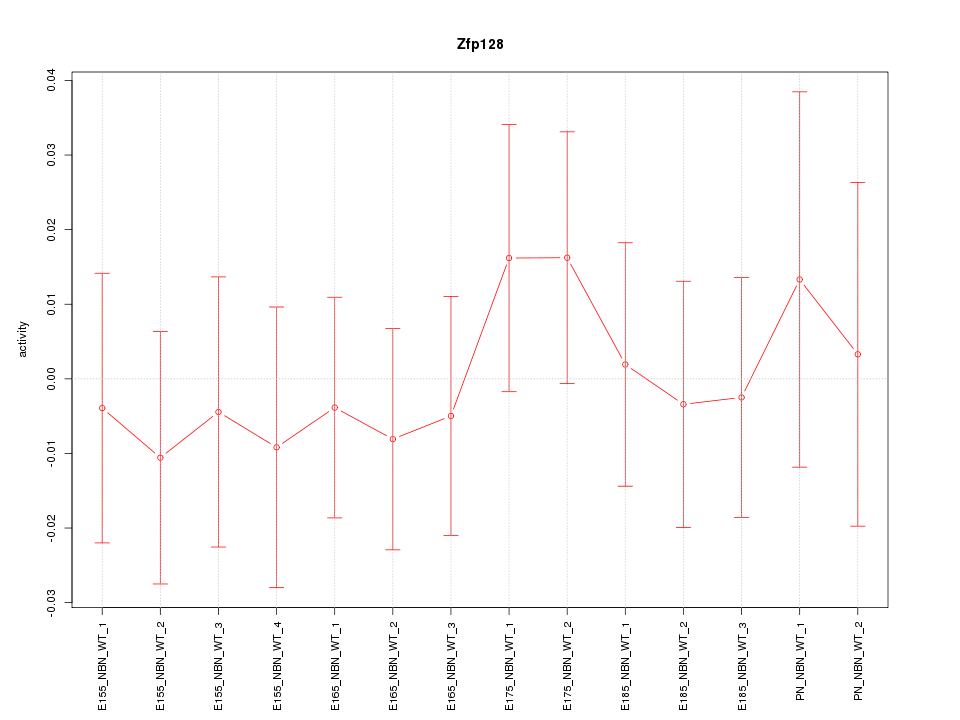
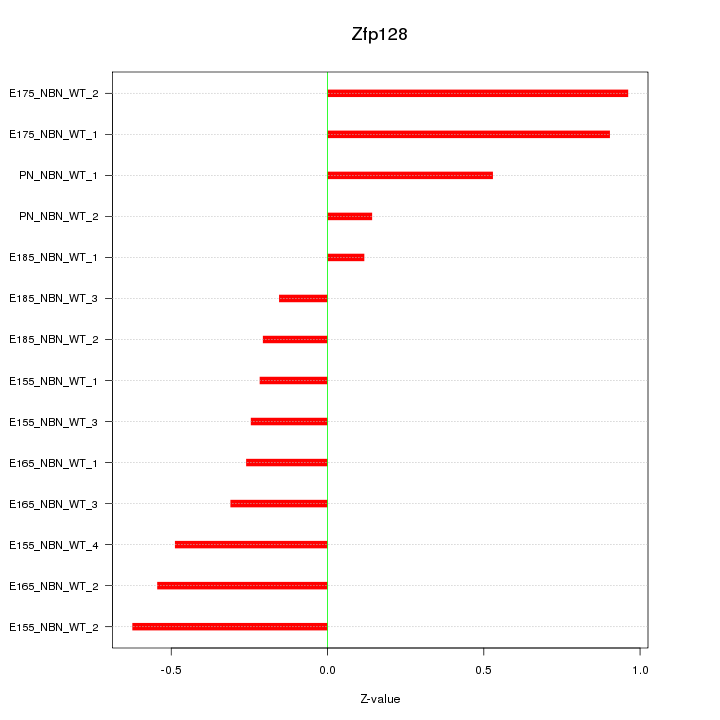
Motif ID: Zfp128
Z-value: 0.487
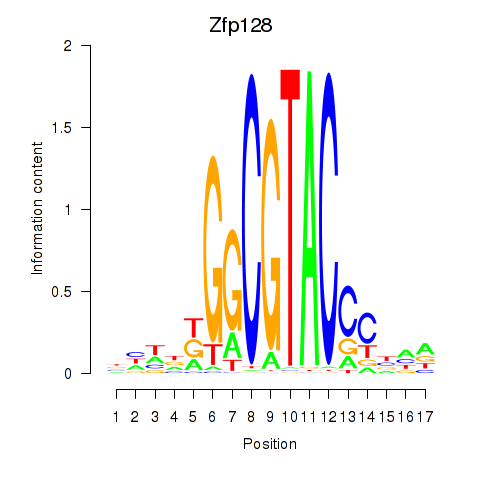
Transcription factors associated with Zfp128:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Zfp128 | ENSMUSG00000060397.6 | Zfp128 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zfp128 | mm10_v2_chr7_+_12881165_12881204 | -0.11 | 7.1e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0090425 | hepatocyte cell migration(GO:0002194) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 0.3 | 0.8 | GO:0043379 | memory T cell differentiation(GO:0043379) |
| 0.2 | 0.7 | GO:0060437 | lung growth(GO:0060437) |
| 0.1 | 0.6 | GO:0060161 | positive regulation of dopamine receptor signaling pathway(GO:0060161) |
| 0.1 | 0.4 | GO:0048597 | post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.0 | 0.6 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.2 | GO:2000781 | positive regulation of double-strand break repair(GO:2000781) |
| 0.0 | 0.2 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 | 0.5 | GO:0007614 | short-term memory(GO:0007614) |
| 0.0 | 0.2 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.0 | GO:0051031 | tRNA transport(GO:0051031) |
| 0.0 | 0.0 | GO:1903546 | microtubule anchoring at centrosome(GO:0034454) negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) protein localization to photoreceptor outer segment(GO:1903546) |
| 0.0 | 0.9 | GO:0008306 | associative learning(GO:0008306) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | GO:0002080 | acrosomal membrane(GO:0002080) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.1 | 0.6 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 0.4 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.8 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 0.7 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.2 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.5 | GO:0031748 | D1 dopamine receptor binding(GO:0031748) |
| 0.0 | 0.1 | GO:0019841 | retinol binding(GO:0019841) |