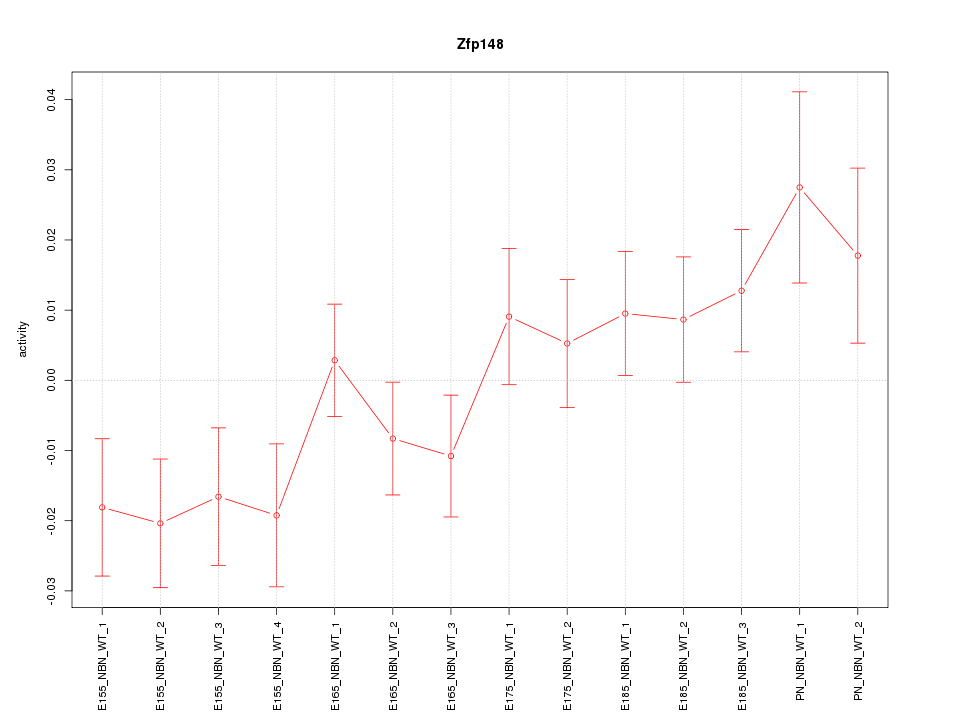
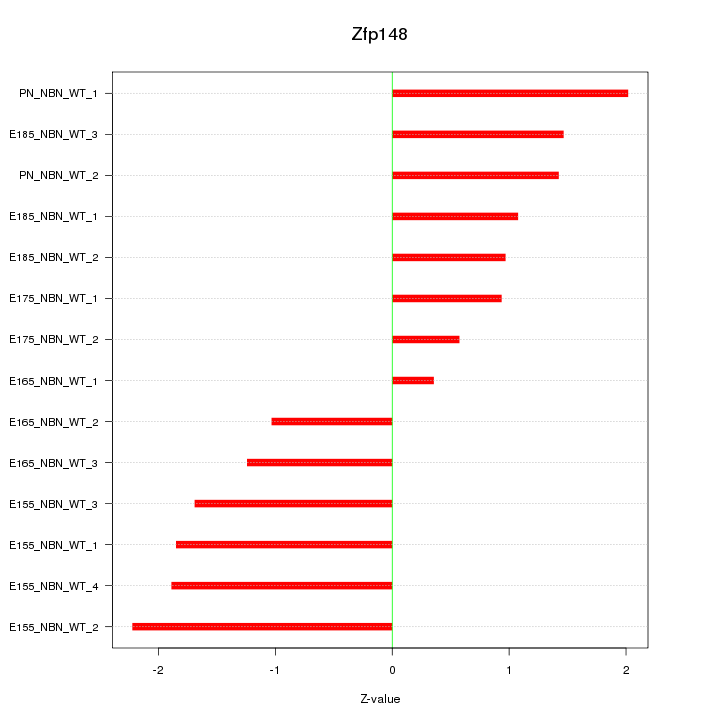
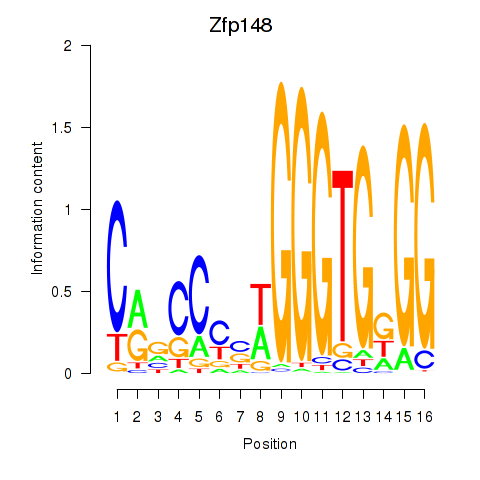
Motif ID: Zfp148
Z-value: 1.442
Transcription factors associated with Zfp148:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Zfp148 | ENSMUSG00000022811.10 | Zfp148 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zfp148 | mm10_v2_chr16_+_33380765_33380787 | 0.08 | 7.8e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 16.3 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 1.9 | 5.6 | GO:1904685 | positive regulation of metalloendopeptidase activity(GO:1904685) |
| 1.5 | 8.7 | GO:0010624 | regulation of Schwann cell proliferation(GO:0010624) |
| 1.1 | 6.9 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 1.1 | 3.3 | GO:0071677 | positive regulation of mononuclear cell migration(GO:0071677) |
| 1.0 | 3.0 | GO:0042939 | renal sodium ion transport(GO:0003096) glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.9 | 2.7 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.6 | 1.9 | GO:0035633 | maintenance of blood-brain barrier(GO:0035633) drug export(GO:0046618) |
| 0.6 | 2.6 | GO:0006842 | tricarboxylic acid transport(GO:0006842) succinate transport(GO:0015744) citrate transport(GO:0015746) |
| 0.6 | 2.4 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.5 | 2.2 | GO:0006867 | asparagine transport(GO:0006867) glutamine transport(GO:0006868) |
| 0.5 | 3.0 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.5 | 1.5 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.5 | 1.9 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.4 | 3.0 | GO:2000598 | regulation of cyclin catabolic process(GO:2000598) negative regulation of cyclin catabolic process(GO:2000599) |
| 0.4 | 0.8 | GO:0021649 | vestibulocochlear nerve morphogenesis(GO:0021648) vestibulocochlear nerve structural organization(GO:0021649) cerebral cortex tangential migration using cell-axon interactions(GO:0021824) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) ganglion morphogenesis(GO:0061552) facioacoustic ganglion development(GO:1903375) dorsal root ganglion morphogenesis(GO:1904835) |
| 0.4 | 1.2 | GO:0006558 | L-phenylalanine metabolic process(GO:0006558) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) |
| 0.4 | 1.1 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.4 | 1.1 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.3 | 1.0 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.3 | 0.9 | GO:0046098 | purine nucleobase catabolic process(GO:0006145) guanine metabolic process(GO:0046098) |
| 0.3 | 0.9 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 0.3 | 2.6 | GO:0032226 | positive regulation of synaptic transmission, dopaminergic(GO:0032226) |
| 0.3 | 2.0 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.3 | 1.0 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.3 | 4.4 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.2 | 1.2 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.2 | 2.4 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.2 | 0.9 | GO:0070589 | cell wall mannoprotein biosynthetic process(GO:0000032) mannoprotein metabolic process(GO:0006056) mannoprotein biosynthetic process(GO:0006057) cell wall glycoprotein biosynthetic process(GO:0031506) cell wall biogenesis(GO:0042546) cell wall macromolecule metabolic process(GO:0044036) cell wall macromolecule biosynthetic process(GO:0044038) chain elongation of O-linked mannose residue(GO:0044845) cellular component macromolecule biosynthetic process(GO:0070589) cell wall organization or biogenesis(GO:0071554) |
| 0.2 | 0.9 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.2 | 0.8 | GO:1900740 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.2 | 1.3 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.2 | 0.8 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.2 | 1.2 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.2 | 1.6 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.2 | 1.2 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.2 | 2.6 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.2 | 1.0 | GO:2000668 | dendritic cell apoptotic process(GO:0097048) regulation of dendritic cell apoptotic process(GO:2000668) |
| 0.2 | 1.9 | GO:0060346 | bone trabecula formation(GO:0060346) |
| 0.2 | 1.1 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.2 | 0.5 | GO:1903061 | regulation of protein lipidation(GO:1903059) positive regulation of protein lipidation(GO:1903061) |
| 0.2 | 0.8 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.2 | 1.5 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.2 | 1.7 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.2 | 0.5 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.1 | 0.4 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.1 | 0.4 | GO:0061153 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.1 | 0.6 | GO:0033092 | positive regulation of immature T cell proliferation in thymus(GO:0033092) |
| 0.1 | 0.8 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.1 | 0.4 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.1 | 0.5 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.1 | 0.5 | GO:0035521 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.1 | 0.7 | GO:0007343 | egg activation(GO:0007343) |
| 0.1 | 2.6 | GO:0019430 | removal of superoxide radicals(GO:0019430) |
| 0.1 | 0.5 | GO:0097114 | NMDA glutamate receptor clustering(GO:0097114) |
| 0.1 | 1.3 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 1.9 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.1 | 1.7 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.1 | 1.3 | GO:0042403 | thyroid hormone metabolic process(GO:0042403) |
| 0.1 | 1.3 | GO:0095500 | acetylcholine receptor signaling pathway(GO:0095500) postsynaptic signal transduction(GO:0098926) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.1 | 0.3 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.1 | 1.1 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 0.1 | 1.1 | GO:0051103 | DNA ligation involved in DNA repair(GO:0051103) |
| 0.1 | 0.6 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.1 | 0.8 | GO:0070813 | hydrogen sulfide metabolic process(GO:0070813) |
| 0.1 | 0.4 | GO:0032627 | interleukin-23 production(GO:0032627) regulation of interleukin-23 production(GO:0032667) |
| 0.1 | 0.3 | GO:1904154 | protein localization to endoplasmic reticulum exit site(GO:0070973) positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.1 | 0.3 | GO:0010757 | arachidonic acid metabolite production involved in inflammatory response(GO:0002538) negative regulation of plasminogen activation(GO:0010757) regulation of vascular wound healing(GO:0061043) regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.1 | 0.5 | GO:1900186 | negative regulation of clathrin-mediated endocytosis(GO:1900186) regulation of caveolin-mediated endocytosis(GO:2001286) |
| 0.1 | 0.5 | GO:0097343 | ripoptosome assembly(GO:0097343) ripoptosome assembly involved in necroptotic process(GO:1901026) |
| 0.1 | 0.4 | GO:0060178 | regulation of exocyst localization(GO:0060178) |
| 0.1 | 1.1 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 0.8 | GO:1901660 | calcium ion export(GO:1901660) |
| 0.1 | 0.6 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.1 | 0.4 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.1 | 0.9 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.1 | 0.3 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.1 | 4.2 | GO:0048747 | muscle fiber development(GO:0048747) |
| 0.1 | 1.1 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.1 | 0.5 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.1 | 0.6 | GO:1901339 | regulation of store-operated calcium channel activity(GO:1901339) |
| 0.1 | 0.2 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.1 | 0.4 | GO:0033089 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.1 | 0.7 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.1 | 1.3 | GO:1900027 | regulation of ruffle assembly(GO:1900027) |
| 0.1 | 0.4 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 0.9 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.0 | 0.3 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.0 | 0.4 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 1.9 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.4 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 1.8 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.2 | GO:0002035 | brain renin-angiotensin system(GO:0002035) |
| 0.0 | 0.3 | GO:1900086 | regulation of peptidyl-tyrosine autophosphorylation(GO:1900084) positive regulation of peptidyl-tyrosine autophosphorylation(GO:1900086) |
| 0.0 | 0.5 | GO:0000076 | DNA replication checkpoint(GO:0000076) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.9 | GO:0040034 | regulation of development, heterochronic(GO:0040034) |
| 0.0 | 0.8 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.1 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.0 | 0.2 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.0 | 0.1 | GO:0002408 | myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.0 | 0.2 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.4 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.5 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.0 | 0.4 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 1.5 | GO:0042632 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.0 | 0.8 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.6 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.1 | GO:0010668 | ectodermal cell differentiation(GO:0010668) chorio-allantoic fusion(GO:0060710) |
| 0.0 | 1.0 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.2 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.0 | 1.0 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.2 | GO:0016446 | somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 | 0.1 | GO:0060339 | negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 0.0 | 1.0 | GO:0015908 | fatty acid transport(GO:0015908) |
| 0.0 | 0.3 | GO:0046855 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 | 0.1 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.3 | GO:0003416 | endochondral bone growth(GO:0003416) |
| 0.0 | 0.6 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) |
| 0.0 | 0.6 | GO:0048255 | mRNA stabilization(GO:0048255) |
| 0.0 | 0.4 | GO:0060441 | epithelial tube branching involved in lung morphogenesis(GO:0060441) |
| 0.0 | 0.2 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.1 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) trophectodermal cell proliferation(GO:0001834) |
| 0.0 | 0.2 | GO:0060324 | face development(GO:0060324) |
| 0.0 | 0.2 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 1.3 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.0 | 0.9 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.2 | GO:0031268 | pseudopodium organization(GO:0031268) |
| 0.0 | 0.6 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.2 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.2 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 0.2 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.0 | 0.5 | GO:0021696 | cerebellar cortex morphogenesis(GO:0021696) |
| 0.0 | 0.3 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.0 | 0.2 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 6.9 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 2.2 | 8.7 | GO:0097450 | astrocyte end-foot(GO:0097450) |
| 1.0 | 3.0 | GO:0031021 | interphase microtubule organizing center(GO:0031021) |
| 0.6 | 3.0 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.4 | 1.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.2 | 0.9 | GO:0031502 | dolichyl-phosphate-mannose-protein mannosyltransferase complex(GO:0031502) |
| 0.2 | 1.7 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.2 | 1.7 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.2 | 1.1 | GO:0008091 | spectrin(GO:0008091) |
| 0.2 | 0.9 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.2 | 1.1 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.2 | 0.7 | GO:0032021 | NELF complex(GO:0032021) |
| 0.2 | 0.9 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.1 | 0.4 | GO:0033193 | Lsd1/2 complex(GO:0033193) |
| 0.1 | 1.4 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.1 | 1.9 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.1 | 0.4 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.1 | 1.2 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.1 | 2.6 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 0.3 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.1 | 0.5 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.1 | 0.8 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 1.1 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.1 | 0.4 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 0.9 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.1 | 0.9 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.1 | 0.5 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.1 | 0.3 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.1 | 0.3 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.1 | 0.4 | GO:0060091 | kinocilium(GO:0060091) |
| 0.1 | 0.6 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.1 | 0.2 | GO:0005940 | septin ring(GO:0005940) |
| 0.1 | 0.2 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.0 | 0.5 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 1.0 | GO:0043657 | host(GO:0018995) host cell part(GO:0033643) host cell(GO:0043657) |
| 0.0 | 4.2 | GO:0016528 | sarcoplasm(GO:0016528) |
| 0.0 | 0.1 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.0 | 0.3 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.2 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.0 | 5.0 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 1.0 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 0.7 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.5 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 0.3 | GO:0097342 | death-inducing signaling complex(GO:0031264) ripoptosome(GO:0097342) |
| 0.0 | 1.3 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 0.2 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 0.3 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 1.0 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 2.0 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.6 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.5 | GO:0008305 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 1.3 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.4 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.7 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.4 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 1.3 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.8 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 1.0 | GO:0030016 | myofibril(GO:0030016) |
| 0.0 | 2.2 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.1 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.9 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.6 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.1 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.0 | 1.9 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.6 | GO:0000791 | euchromatin(GO:0000791) |
| 0.0 | 0.9 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.6 | GO:0044439 | microbody part(GO:0044438) peroxisomal part(GO:0044439) |
| 0.0 | 0.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.2 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.5 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 5.7 | GO:0005912 | adherens junction(GO:0005912) |
| 0.0 | 8.9 | GO:0005615 | extracellular space(GO:0005615) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 16.1 | GO:0038191 | neuropilin binding(GO:0038191) |
| 1.0 | 6.9 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.9 | 2.6 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.6 | 5.6 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.5 | 2.0 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.4 | 2.2 | GO:0015186 | L-asparagine transmembrane transporter activity(GO:0015182) L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.4 | 2.4 | GO:0002054 | nucleobase binding(GO:0002054) |
| 0.4 | 1.9 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.4 | 1.1 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) bisphosphoglycerate 2-phosphatase activity(GO:0004083) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.3 | 1.0 | GO:0019002 | GMP binding(GO:0019002) |
| 0.3 | 3.0 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.3 | 1.2 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.3 | 1.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.3 | 1.3 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.3 | 1.0 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.2 | 5.4 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.2 | 2.6 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.2 | 2.7 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.2 | 0.6 | GO:0043758 | acetate-CoA ligase (ADP-forming) activity(GO:0043758) |
| 0.2 | 0.7 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.2 | 1.1 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.2 | 0.2 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.2 | 0.5 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.2 | 0.7 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.2 | 1.1 | GO:0045703 | pinocarveol dehydrogenase activity(GO:0018446) chloral hydrate dehydrogenase activity(GO:0018447) hydroxymethylmethylsilanediol oxidase activity(GO:0018448) 1-phenylethanol dehydrogenase activity(GO:0018449) myrtenol dehydrogenase activity(GO:0018450) cis-1,2-dihydroxy-1,2-dihydro-8-carboxynaphthalene dehydrogenase activity(GO:0034522) 3-hydroxy-4-methyloctanoyl-CoA dehydrogenase activity(GO:0034582) 2-hydroxy-4-isopropenylcyclohexane-1-carboxyl-CoA dehydrogenase activity(GO:0034778) cis-9,10-dihydroanthracene-9,10-diol dehydrogenase activity(GO:0034817) citronellol dehydrogenase activity(GO:0034821) naphthyl-2-hydroxymethyl-succinyl-CoA dehydrogenase activity(GO:0034847) 2,4,4-trimethyl-1-pentanol dehydrogenase activity(GO:0034863) 2,4,4-trimethyl-3-hydroxypentanoyl-CoA dehydrogenase activity(GO:0034868) 1-hydroxy-4,4-dimethylpentan-3-one dehydrogenase activity(GO:0034871) endosulfan diol dehydrogenase activity(GO:0034891) endosulfan hydroxyether dehydrogenase activity(GO:0034901) 3-hydroxy-2-methylhexanoyl-CoA dehydrogenase activity(GO:0034918) 3-hydroxy-2,6-dimethyl-5-methylene-heptanoyl-CoA dehydrogenase activity(GO:0034944) versicolorin reductase activity(GO:0042469) ketoreductase activity(GO:0045703) |
| 0.2 | 1.3 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.2 | 4.4 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.2 | 0.3 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.1 | 9.4 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 0.9 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 0.6 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 1.7 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.1 | 0.5 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 1.3 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 0.8 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.1 | 3.4 | GO:0042805 | actinin binding(GO:0042805) |
| 0.1 | 2.4 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 1.0 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 0.5 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.1 | 1.5 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 1.9 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 0.4 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.1 | 0.8 | GO:0016701 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen(GO:0016701) oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.1 | 3.0 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.1 | 0.5 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.1 | 0.7 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 0.3 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.1 | 0.4 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.1 | 0.3 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.1 | 3.0 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 1.4 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 1.4 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 0.6 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.1 | 0.7 | GO:0034943 | acyl-CoA ligase activity(GO:0003996) 3-oxo-2-(2'-pentenyl)cyclopentane-1-octanoic acid CoA ligase activity(GO:0010435) 3-isopropenyl-6-oxoheptanoyl-CoA synthetase activity(GO:0018854) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA synthetase activity(GO:0018855) benzoyl acetate-CoA ligase activity(GO:0018856) 2,4-dichlorobenzoate-CoA ligase activity(GO:0018857) very long-chain fatty acid-CoA ligase activity(GO:0031957) pivalate-CoA ligase activity(GO:0034783) cyclopropanecarboxylate-CoA ligase activity(GO:0034793) adipate-CoA ligase activity(GO:0034796) citronellyl-CoA ligase activity(GO:0034823) mentha-1,3-dione-CoA ligase activity(GO:0034841) thiophene-2-carboxylate-CoA ligase activity(GO:0034842) 2,4,4-trimethylpentanoate-CoA ligase activity(GO:0034865) cis-2-methyl-5-isopropylhexa-2,5-dienoate-CoA ligase activity(GO:0034942) trans-2-methyl-5-isopropylhexa-2,5-dienoate-CoA ligase activity(GO:0034943) branched-chain acyl-CoA synthetase (ADP-forming) activity(GO:0043759) aryl-CoA synthetase (ADP-forming) activity(GO:0043762) 3-hydroxypropionyl-CoA synthetase activity(GO:0043955) perillic acid:CoA ligase (ADP-forming) activity(GO:0052685) perillic acid:CoA ligase (AMP-forming) activity(GO:0052686) (3R)-3-isopropenyl-6-oxoheptanoate:CoA ligase (ADP-forming) activity(GO:0052687) (3R)-3-isopropenyl-6-oxoheptanoate:CoA ligase (AMP-forming) activity(GO:0052688) pristanate-CoA ligase activity(GO:0070251) malonyl-CoA synthetase activity(GO:0090409) |
| 0.1 | 0.7 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.1 | 0.4 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 0.8 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 0.9 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.1 | 0.2 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 0.2 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.1 | 1.2 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 0.2 | GO:0008297 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) double-stranded DNA exodeoxyribonuclease activity(GO:0008309) |
| 0.1 | 0.5 | GO:0016920 | pyroglutamyl-peptidase activity(GO:0016920) |
| 0.1 | 1.3 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.5 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.0 | 0.5 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.9 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 5.2 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 1.0 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.4 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.4 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.0 | 1.7 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 1.1 | GO:0005272 | sodium channel activity(GO:0005272) |
| 0.0 | 0.3 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.0 | 2.0 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.3 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.5 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.9 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.6 | GO:0015215 | nucleotide transmembrane transporter activity(GO:0015215) |
| 0.0 | 1.2 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.2 | GO:0051718 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates(GO:0051718) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.8 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 1.2 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.9 | GO:0016814 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amidines(GO:0016814) |
| 0.0 | 0.2 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.0 | GO:0051429 | corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.0 | 0.4 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.4 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.4 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 1.6 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.9 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.0 | 0.5 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 1.4 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 2.1 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 2.2 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.4 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.3 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 1.0 | GO:0101005 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.2 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.2 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |