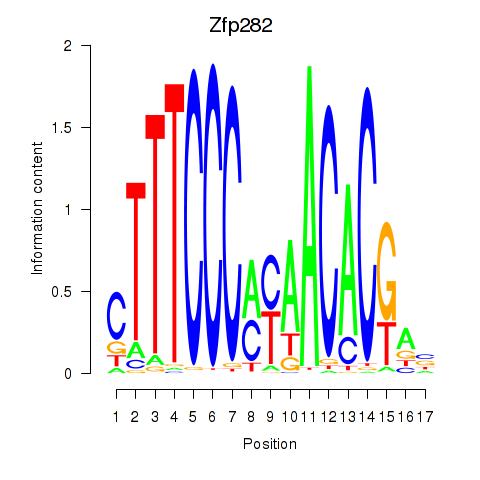
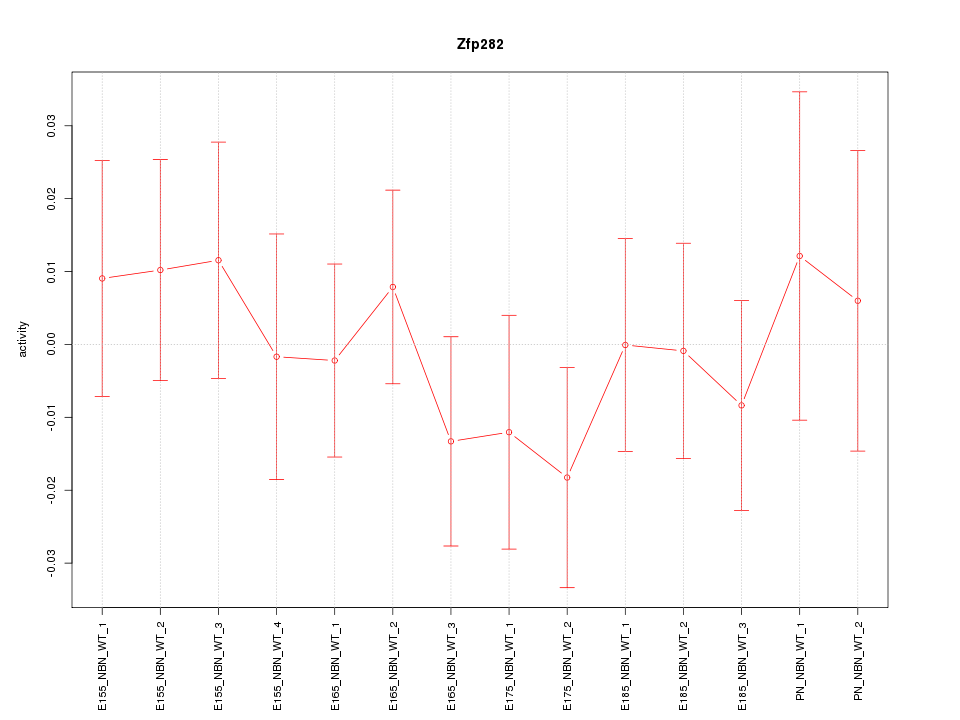
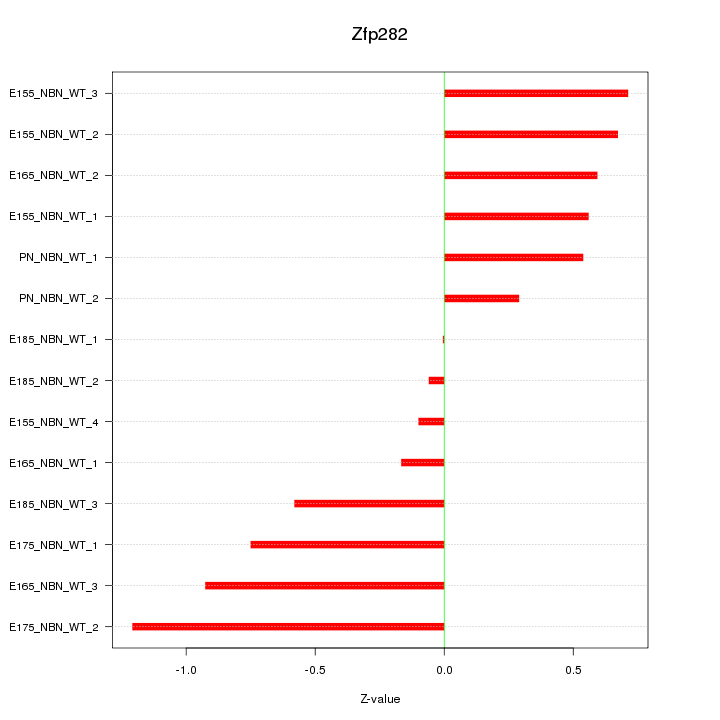
| 0.3 |
0.8 |
GO:0048389 |
limb joint morphogenesis(GO:0036022) embryonic skeletal limb joint morphogenesis(GO:0036023) intermediate mesoderm development(GO:0048389) pattern specification involved in mesonephros development(GO:0061227) anterior/posterior pattern specification involved in kidney development(GO:0072098) mesonephric duct development(GO:0072177) mesonephric duct morphogenesis(GO:0072180) ureter urothelium development(GO:0072190) kidney smooth muscle tissue development(GO:0072194) |
| 0.2 |
0.9 |
GO:0002121 |
inter-male aggressive behavior(GO:0002121) |
| 0.2 |
0.9 |
GO:0046133 |
pyrimidine ribonucleoside catabolic process(GO:0046133) |
| 0.2 |
0.7 |
GO:0002337 |
B-1a B cell differentiation(GO:0002337) |
| 0.2 |
0.5 |
GO:0070488 |
neutrophil aggregation(GO:0070488) |
| 0.1 |
0.4 |
GO:0045404 |
positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.1 |
0.6 |
GO:0002457 |
T cell antigen processing and presentation(GO:0002457) |
| 0.1 |
0.5 |
GO:0036324 |
vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 0.1 |
0.5 |
GO:0097460 |
ferrous iron import into cell(GO:0097460) |
| 0.1 |
0.2 |
GO:0035521 |
monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.1 |
0.2 |
GO:0046709 |
IDP metabolic process(GO:0046707) IDP catabolic process(GO:0046709) |
| 0.1 |
0.8 |
GO:2000353 |
positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.0 |
0.1 |
GO:0009106 |
lipoate metabolic process(GO:0009106) |
| 0.0 |
0.4 |
GO:0048742 |
regulation of skeletal muscle fiber development(GO:0048742) |
| 0.0 |
0.6 |
GO:0033327 |
Leydig cell differentiation(GO:0033327) |
| 0.0 |
0.3 |
GO:0016338 |
calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 |
0.5 |
GO:0046697 |
decidualization(GO:0046697) |
| 0.0 |
0.1 |
GO:1903774 |
positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.0 |
0.1 |
GO:0048208 |
vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.0 |
0.1 |
GO:0051611 |
negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 |
0.1 |
GO:0006667 |
sphinganine metabolic process(GO:0006667) |
| 0.0 |
0.3 |
GO:2000010 |
positive regulation of protein localization to cell surface(GO:2000010) |
| 0.0 |
0.3 |
GO:0042347 |
negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.0 |
0.1 |
GO:0098535 |
de novo centriole assembly(GO:0098535) |
| 0.0 |
0.1 |
GO:0006703 |
estrogen biosynthetic process(GO:0006703) |
| 0.0 |
0.2 |
GO:0009083 |
branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 |
0.2 |
GO:0019363 |
pyridine nucleotide biosynthetic process(GO:0019363) |