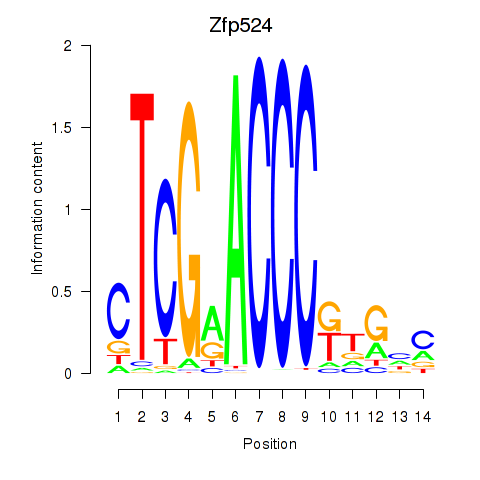
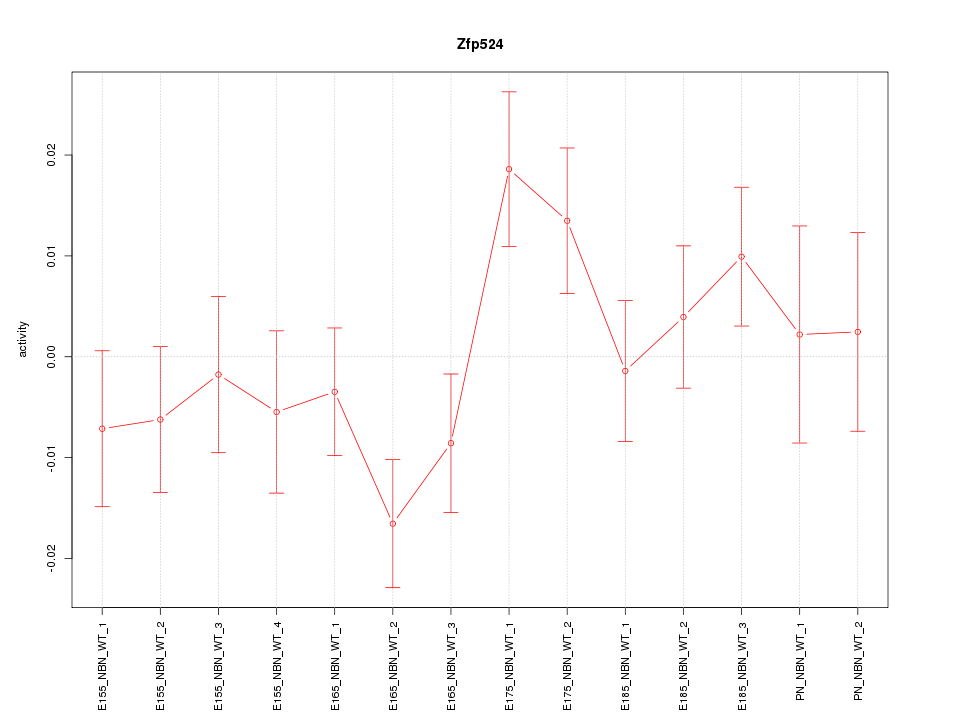
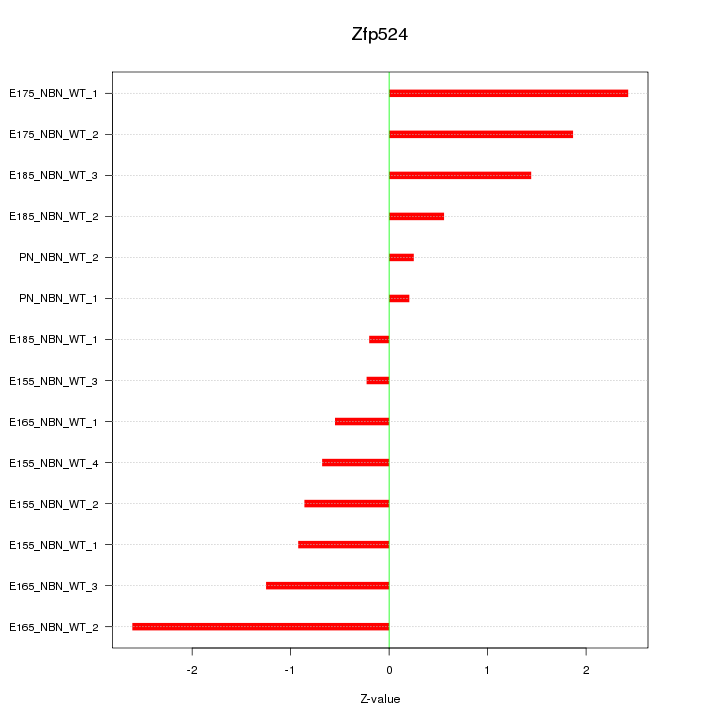
| 1.0 |
3.9 |
GO:0035624 |
receptor transactivation(GO:0035624) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) |
| 0.6 |
3.0 |
GO:2000065 |
negative regulation of aldosterone metabolic process(GO:0032345) negative regulation of aldosterone biosynthetic process(GO:0032348) negative regulation of cortisol biosynthetic process(GO:2000065) |
| 0.5 |
1.4 |
GO:0032430 |
positive regulation of phospholipase A2 activity(GO:0032430) |
| 0.4 |
1.8 |
GO:0032423 |
regulation of mismatch repair(GO:0032423) |
| 0.3 |
1.0 |
GO:0021837 |
motogenic signaling involved in postnatal olfactory bulb interneuron migration(GO:0021837) positive regulation of mitotic cell cycle DNA replication(GO:1903465) |
| 0.3 |
0.6 |
GO:0000083 |
regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.3 |
1.7 |
GO:0050861 |
positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.3 |
1.4 |
GO:0015788 |
UDP-N-acetylglucosamine transport(GO:0015788) |
| 0.3 |
0.9 |
GO:0051542 |
elastin biosynthetic process(GO:0051542) |
| 0.2 |
0.6 |
GO:0071492 |
cellular response to UV-A(GO:0071492) |
| 0.2 |
0.6 |
GO:0042196 |
dichloromethane metabolic process(GO:0018900) chlorinated hydrocarbon metabolic process(GO:0042196) halogenated hydrocarbon metabolic process(GO:0042197) |
| 0.2 |
4.8 |
GO:2000310 |
regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.2 |
1.1 |
GO:0045218 |
zonula adherens maintenance(GO:0045218) |
| 0.2 |
0.7 |
GO:0098915 |
membrane repolarization during ventricular cardiac muscle cell action potential(GO:0098915) |
| 0.2 |
0.5 |
GO:0001866 |
NK T cell proliferation(GO:0001866) |
| 0.2 |
0.5 |
GO:0009193 |
pyrimidine ribonucleoside diphosphate metabolic process(GO:0009193) UDP metabolic process(GO:0046048) |
| 0.2 |
0.5 |
GO:0036091 |
positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.2 |
0.8 |
GO:0010032 |
meiotic chromosome condensation(GO:0010032) |
| 0.2 |
0.5 |
GO:2000987 |
positive regulation of fear response(GO:1903367) positive regulation of behavioral fear response(GO:2000987) |
| 0.1 |
1.7 |
GO:0032688 |
negative regulation of interferon-beta production(GO:0032688) |
| 0.1 |
4.1 |
GO:0008299 |
isoprenoid biosynthetic process(GO:0008299) |
| 0.1 |
0.6 |
GO:0010700 |
negative regulation of norepinephrine secretion(GO:0010700) |
| 0.1 |
0.4 |
GO:0032066 |
nucleolus to nucleoplasm transport(GO:0032066) |
| 0.1 |
0.7 |
GO:0006543 |
glutamine catabolic process(GO:0006543) |
| 0.1 |
0.6 |
GO:1903361 |
protein localization to basolateral plasma membrane(GO:1903361) |
| 0.1 |
0.5 |
GO:0046950 |
cellular ketone body metabolic process(GO:0046950) |
| 0.1 |
1.2 |
GO:1903608 |
protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.1 |
0.7 |
GO:0051694 |
pointed-end actin filament capping(GO:0051694) |
| 0.1 |
0.8 |
GO:0043084 |
penile erection(GO:0043084) |
| 0.1 |
0.5 |
GO:0031622 |
positive regulation of fever generation(GO:0031622) |
| 0.1 |
0.6 |
GO:1900122 |
positive regulation of receptor binding(GO:1900122) |
| 0.1 |
0.3 |
GO:0002636 |
positive regulation of germinal center formation(GO:0002636) |
| 0.1 |
0.4 |
GO:1901301 |
regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.1 |
1.6 |
GO:0002495 |
antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.1 |
0.6 |
GO:0003383 |
apical constriction(GO:0003383) |
| 0.1 |
1.1 |
GO:0048484 |
enteric nervous system development(GO:0048484) |
| 0.1 |
0.5 |
GO:0032713 |
negative regulation of interleukin-4 production(GO:0032713) |
| 0.1 |
0.5 |
GO:0032911 |
negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.1 |
0.4 |
GO:0030242 |
pexophagy(GO:0030242) |
| 0.1 |
0.3 |
GO:0010512 |
negative regulation of phosphatidylinositol biosynthetic process(GO:0010512) |
| 0.1 |
1.6 |
GO:0006958 |
complement activation, classical pathway(GO:0006958) |
| 0.1 |
0.6 |
GO:0006620 |
posttranslational protein targeting to membrane(GO:0006620) |
| 0.1 |
0.2 |
GO:0009174 |
UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.1 |
0.4 |
GO:0090148 |
membrane fission(GO:0090148) |
| 0.1 |
0.5 |
GO:0015808 |
L-alanine transport(GO:0015808) |
| 0.1 |
0.2 |
GO:0002331 |
pre-B cell allelic exclusion(GO:0002331) signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.1 |
0.4 |
GO:0042590 |
antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.1 |
0.7 |
GO:0009404 |
toxin metabolic process(GO:0009404) |
| 0.1 |
0.3 |
GO:0035521 |
monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.1 |
0.2 |
GO:0010533 |
regulation of activation of Janus kinase activity(GO:0010533) regulation of activation of JAK2 kinase activity(GO:0010534) |
| 0.1 |
0.5 |
GO:0071318 |
cellular response to ATP(GO:0071318) |
| 0.1 |
0.3 |
GO:1901252 |
regulation of intracellular transport of viral material(GO:1901252) |
| 0.1 |
0.3 |
GO:0072383 |
plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.1 |
1.7 |
GO:0032011 |
ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 |
0.3 |
GO:0051890 |
regulation of cardioblast differentiation(GO:0051890) |
| 0.1 |
0.3 |
GO:0006049 |
UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.1 |
1.7 |
GO:0001573 |
ganglioside metabolic process(GO:0001573) |
| 0.1 |
0.3 |
GO:1904694 |
negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.1 |
0.2 |
GO:0009051 |
pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.1 |
0.2 |
GO:0031581 |
hemidesmosome assembly(GO:0031581) |
| 0.1 |
0.2 |
GO:0042732 |
D-xylose metabolic process(GO:0042732) |
| 0.1 |
0.3 |
GO:0007144 |
female meiosis I(GO:0007144) |
| 0.1 |
0.1 |
GO:0006295 |
nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.1 |
0.3 |
GO:0034316 |
negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.1 |
0.5 |
GO:0071420 |
cellular response to histamine(GO:0071420) |
| 0.1 |
0.6 |
GO:0097011 |
cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) |
| 0.1 |
0.8 |
GO:0042759 |
long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.1 |
0.6 |
GO:0051307 |
meiotic chromosome separation(GO:0051307) |
| 0.1 |
0.2 |
GO:0051031 |
tRNA transport(GO:0051031) |
| 0.1 |
0.6 |
GO:0090557 |
establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 |
0.5 |
GO:0007042 |
lysosomal lumen acidification(GO:0007042) |
| 0.1 |
0.3 |
GO:0035520 |
monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 |
0.2 |
GO:1902255 |
cerebrospinal fluid secretion(GO:0033326) positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902255) |
| 0.0 |
0.1 |
GO:0071929 |
alpha-tubulin acetylation(GO:0071929) |
| 0.0 |
0.1 |
GO:2001286 |
regulation of caveolin-mediated endocytosis(GO:2001286) |
| 0.0 |
0.2 |
GO:0090035 |
regulation of chaperone-mediated protein complex assembly(GO:0090034) positive regulation of chaperone-mediated protein complex assembly(GO:0090035) |
| 0.0 |
0.4 |
GO:2000051 |
negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.0 |
0.6 |
GO:0072010 |
renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.0 |
0.8 |
GO:0043552 |
positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.0 |
0.2 |
GO:0061502 |
early endosome to recycling endosome transport(GO:0061502) |
| 0.0 |
0.3 |
GO:0072321 |
chaperone-mediated protein transport(GO:0072321) |
| 0.0 |
0.6 |
GO:0043249 |
erythrocyte maturation(GO:0043249) |
| 0.0 |
0.2 |
GO:0034454 |
microtubule anchoring at centrosome(GO:0034454) |
| 0.0 |
0.2 |
GO:0042795 |
snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 |
0.4 |
GO:0000290 |
deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 |
0.6 |
GO:0036093 |
germ cell proliferation(GO:0036093) |
| 0.0 |
0.2 |
GO:0060158 |
phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 |
0.1 |
GO:0086046 |
membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.0 |
0.1 |
GO:0006610 |
ribosomal protein import into nucleus(GO:0006610) |
| 0.0 |
0.4 |
GO:0042996 |
regulation of Golgi to plasma membrane protein transport(GO:0042996) |
| 0.0 |
0.5 |
GO:0006622 |
protein targeting to lysosome(GO:0006622) |
| 0.0 |
0.1 |
GO:0060800 |
regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 |
0.2 |
GO:0006290 |
pyrimidine dimer repair(GO:0006290) |
| 0.0 |
0.4 |
GO:0097067 |
cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 |
0.3 |
GO:0010587 |
miRNA catabolic process(GO:0010587) |
| 0.0 |
0.2 |
GO:0030397 |
membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.0 |
0.1 |
GO:0008295 |
spermidine biosynthetic process(GO:0008295) |
| 0.0 |
0.8 |
GO:0010107 |
potassium ion import(GO:0010107) |
| 0.0 |
0.2 |
GO:0097680 |
double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.0 |
0.2 |
GO:0033129 |
positive regulation of histone phosphorylation(GO:0033129) |
| 0.0 |
0.1 |
GO:0050862 |
positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.0 |
0.4 |
GO:0021942 |
radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.0 |
0.6 |
GO:0036342 |
post-anal tail morphogenesis(GO:0036342) |
| 0.0 |
0.2 |
GO:0018344 |
protein geranylgeranylation(GO:0018344) |
| 0.0 |
0.1 |
GO:2000298 |
regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.0 |
0.1 |
GO:0035973 |
aggrephagy(GO:0035973) |
| 0.0 |
0.2 |
GO:0071442 |
positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.0 |
0.1 |
GO:0071894 |
histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.0 |
0.1 |
GO:0030208 |
dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 |
0.2 |
GO:0035881 |
amacrine cell differentiation(GO:0035881) |
| 0.0 |
0.1 |
GO:1900748 |
positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 |
0.2 |
GO:1903352 |
ornithine transport(GO:0015822) L-ornithine transmembrane transport(GO:1903352) |
| 0.0 |
0.4 |
GO:0048240 |
sperm capacitation(GO:0048240) |
| 0.0 |
0.1 |
GO:0019085 |
early viral transcription(GO:0019085) |
| 0.0 |
0.1 |
GO:1904749 |
regulation of protein localization to nucleolus(GO:1904749) positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.0 |
0.3 |
GO:0033574 |
response to testosterone(GO:0033574) |
| 0.0 |
0.1 |
GO:0019236 |
response to pheromone(GO:0019236) |
| 0.0 |
0.2 |
GO:0097091 |
synaptic vesicle clustering(GO:0097091) |
| 0.0 |
0.3 |
GO:0051382 |
kinetochore assembly(GO:0051382) |
| 0.0 |
0.1 |
GO:0032485 |
regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 |
0.5 |
GO:0008209 |
androgen metabolic process(GO:0008209) |
| 0.0 |
0.0 |
GO:1901897 |
regulation of relaxation of cardiac muscle(GO:1901897) |
| 0.0 |
0.2 |
GO:0006013 |
mannose metabolic process(GO:0006013) |
| 0.0 |
0.4 |
GO:0045292 |
mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 |
0.1 |
GO:0090399 |
replicative senescence(GO:0090399) |
| 0.0 |
0.1 |
GO:0051088 |
PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.0 |
0.3 |
GO:0043923 |
positive regulation by host of viral transcription(GO:0043923) |
| 0.0 |
0.2 |
GO:0045056 |
transcytosis(GO:0045056) |
| 0.0 |
0.3 |
GO:0043162 |
ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 |
1.1 |
GO:0071347 |
cellular response to interleukin-1(GO:0071347) |
| 0.0 |
0.1 |
GO:0051697 |
protein delipidation(GO:0051697) |
| 0.0 |
0.2 |
GO:0031274 |
positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 |
0.3 |
GO:0071364 |
cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.0 |
0.1 |
GO:0010606 |
positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 |
0.7 |
GO:0051703 |
social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.0 |
0.1 |
GO:0007262 |
STAT protein import into nucleus(GO:0007262) |
| 0.0 |
0.1 |
GO:0031133 |
cellular magnesium ion homeostasis(GO:0010961) regulation of axon diameter(GO:0031133) |
| 0.0 |
0.3 |
GO:0006750 |
glutathione biosynthetic process(GO:0006750) |
| 0.0 |
0.6 |
GO:0001945 |
lymph vessel development(GO:0001945) |
| 0.0 |
0.9 |
GO:0008306 |
associative learning(GO:0008306) |
| 0.0 |
0.5 |
GO:0001964 |
startle response(GO:0001964) |
| 0.0 |
0.1 |
GO:0060627 |
regulation of vesicle-mediated transport(GO:0060627) |
| 0.0 |
0.0 |
GO:2000832 |
protein-chromophore linkage(GO:0018298) negative regulation of steroid hormone secretion(GO:2000832) negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.0 |
0.1 |
GO:0045988 |
tachykinin receptor signaling pathway(GO:0007217) negative regulation of striated muscle contraction(GO:0045988) |
| 0.0 |
1.1 |
GO:0050885 |
neuromuscular process controlling balance(GO:0050885) |
| 0.0 |
0.1 |
GO:0006020 |
inositol metabolic process(GO:0006020) |
| 0.0 |
0.2 |
GO:0034244 |
negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 |
0.1 |
GO:0071033 |
nuclear retention of pre-mRNA at the site of transcription(GO:0071033) CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.0 |
1.0 |
GO:0019882 |
antigen processing and presentation(GO:0019882) |
| 0.0 |
0.2 |
GO:0031115 |
negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 |
0.1 |
GO:0032464 |
positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 |
0.1 |
GO:1902626 |
assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 |
0.5 |
GO:0043001 |
Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 |
0.1 |
GO:0032876 |
negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 |
0.0 |
GO:0010626 |
negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.0 |
0.0 |
GO:0000059 |
protein import into nucleus, docking(GO:0000059) |
| 0.0 |
0.2 |
GO:0022027 |
interkinetic nuclear migration(GO:0022027) |
| 0.0 |
0.3 |
GO:0090102 |
cochlea development(GO:0090102) |
| 0.0 |
0.1 |
GO:0006102 |
isocitrate metabolic process(GO:0006102) |
| 0.0 |
0.2 |
GO:0035025 |
positive regulation of Rho protein signal transduction(GO:0035025) |