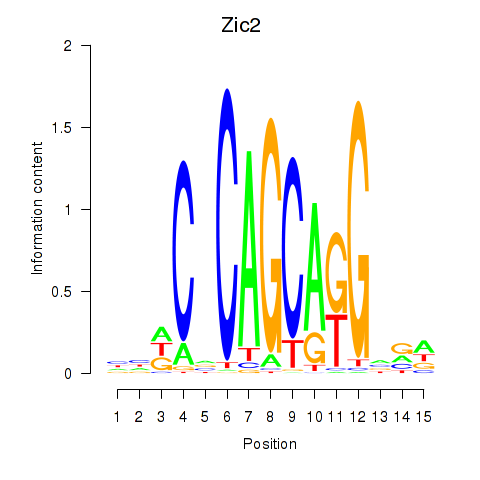
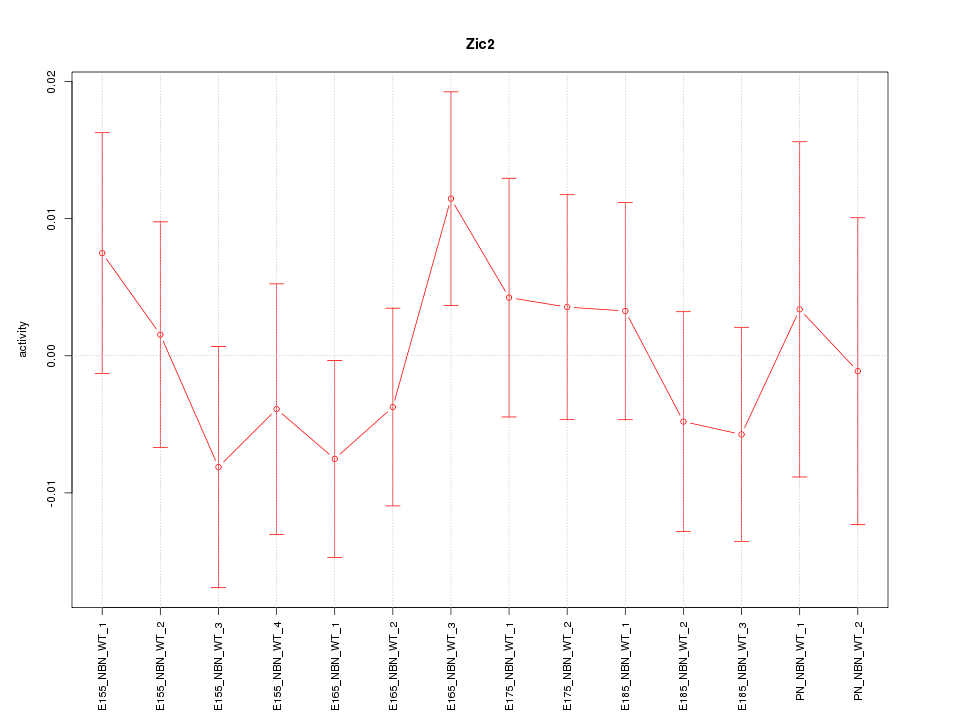
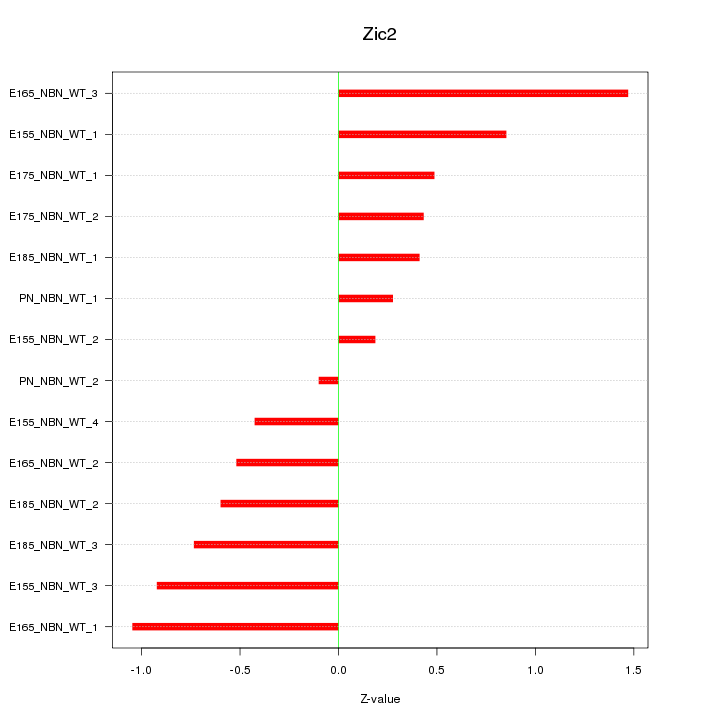
| 0.4 |
1.1 |
GO:2001139 |
negative regulation of postsynaptic membrane organization(GO:1901627) negative regulation of dendritic spine maintenance(GO:1902951) negative regulation of phospholipid efflux(GO:1902999) regulation of lipid transport across blood brain barrier(GO:1903000) negative regulation of lipid transport across blood brain barrier(GO:1903001) positive regulation of lipid transport across blood brain barrier(GO:1903002) negative regulation of phospholipid transport(GO:2001139) |
| 0.2 |
1.2 |
GO:2000255 |
negative regulation of male germ cell proliferation(GO:2000255) |
| 0.2 |
0.5 |
GO:1902358 |
sulfate transmembrane transport(GO:1902358) |
| 0.2 |
0.5 |
GO:0048496 |
maintenance of organ identity(GO:0048496) |
| 0.1 |
0.3 |
GO:1902037 |
positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 0.1 |
0.4 |
GO:0035428 |
hexose transmembrane transport(GO:0035428) |
| 0.1 |
0.2 |
GO:0060126 |
hypophysis morphogenesis(GO:0048850) somatotropin secreting cell differentiation(GO:0060126) |
| 0.1 |
0.2 |
GO:0006116 |
NADH oxidation(GO:0006116) |
| 0.1 |
0.5 |
GO:0061042 |
vascular wound healing(GO:0061042) |
| 0.1 |
0.2 |
GO:0010248 |
B cell negative selection(GO:0002352) establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.1 |
0.5 |
GO:0038028 |
insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.1 |
0.2 |
GO:2000564 |
regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) CD8-positive, alpha-beta T cell proliferation(GO:0035740) negative regulation of regulatory T cell differentiation(GO:0045590) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) |
| 0.0 |
0.1 |
GO:0006601 |
creatine biosynthetic process(GO:0006601) |
| 0.0 |
0.1 |
GO:0032474 |
otolith morphogenesis(GO:0032474) |
| 0.0 |
0.1 |
GO:0035574 |
histone H4-K20 demethylation(GO:0035574) |
| 0.0 |
0.3 |
GO:0016127 |
cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 |
0.2 |
GO:0014012 |
peripheral nervous system axon regeneration(GO:0014012) |
| 0.0 |
0.2 |
GO:0032346 |
positive regulation of aldosterone metabolic process(GO:0032346) positive regulation of aldosterone biosynthetic process(GO:0032349) regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.0 |
0.1 |
GO:0032632 |
interleukin-3 production(GO:0032632) |
| 0.0 |
0.2 |
GO:2000507 |
positive regulation of energy homeostasis(GO:2000507) |
| 0.0 |
0.1 |
GO:0032488 |
Cdc42 protein signal transduction(GO:0032488) |
| 0.0 |
0.1 |
GO:0010756 |
positive regulation of plasminogen activation(GO:0010756) |
| 0.0 |
0.1 |
GO:0010838 |
positive regulation of keratinocyte proliferation(GO:0010838) |
| 0.0 |
0.3 |
GO:0033314 |
mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 |
0.1 |
GO:0048239 |
negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 0.0 |
0.2 |
GO:0051045 |
negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 |
0.2 |
GO:0007386 |
compartment pattern specification(GO:0007386) |
| 0.0 |
0.2 |
GO:0060024 |
rhythmic synaptic transmission(GO:0060024) |
| 0.0 |
0.2 |
GO:0061092 |
regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.0 |
0.2 |
GO:0019695 |
choline metabolic process(GO:0019695) |
| 0.0 |
0.1 |
GO:0002606 |
positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.0 |
0.3 |
GO:1902166 |
negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902166) |
| 0.0 |
0.1 |
GO:0002934 |
desmosome organization(GO:0002934) |
| 0.0 |
0.1 |
GO:1900864 |
mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.0 |
0.2 |
GO:0048251 |
elastic fiber assembly(GO:0048251) |
| 0.0 |
0.1 |
GO:1903998 |
regulation of eating behavior(GO:1903998) |
| 0.0 |
0.1 |
GO:2001015 |
negative regulation of skeletal muscle cell differentiation(GO:2001015) |
| 0.0 |
0.2 |
GO:0046339 |
diacylglycerol metabolic process(GO:0046339) |
| 0.0 |
0.1 |
GO:0043490 |
malate-aspartate shuttle(GO:0043490) |
| 0.0 |
0.1 |
GO:0046959 |
habituation(GO:0046959) |
| 0.0 |
0.1 |
GO:0030576 |
Cajal body organization(GO:0030576) |
| 0.0 |
0.1 |
GO:0086042 |
cardiac muscle cell-cardiac muscle cell adhesion(GO:0086042) |
| 0.0 |
0.1 |
GO:0009405 |
pathogenesis(GO:0009405) |
| 0.0 |
0.1 |
GO:0070829 |
heterochromatin maintenance(GO:0070829) |
| 0.0 |
0.4 |
GO:0032060 |
bleb assembly(GO:0032060) |
| 0.0 |
0.1 |
GO:0003032 |
detection of oxygen(GO:0003032) cell migration involved in endocardial cushion formation(GO:0003273) |
| 0.0 |
0.2 |
GO:0015884 |
folic acid transport(GO:0015884) |
| 0.0 |
0.1 |
GO:0007039 |
protein catabolic process in the vacuole(GO:0007039) |
| 0.0 |
0.2 |
GO:0051775 |
response to redox state(GO:0051775) |
| 0.0 |
0.2 |
GO:0035814 |
negative regulation of renal sodium excretion(GO:0035814) |
| 0.0 |
0.2 |
GO:0000712 |
resolution of meiotic recombination intermediates(GO:0000712) |
| 0.0 |
0.1 |
GO:0030382 |
sperm mitochondrion organization(GO:0030382) |
| 0.0 |
0.1 |
GO:0051771 |
negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.0 |
0.1 |
GO:0070305 |
response to cGMP(GO:0070305) cellular response to cGMP(GO:0071321) |
| 0.0 |
0.0 |
GO:0043181 |
vacuolar sequestering(GO:0043181) |
| 0.0 |
0.0 |
GO:0019482 |
beta-alanine metabolic process(GO:0019482) |
| 0.0 |
0.0 |
GO:0018298 |
protein-chromophore linkage(GO:0018298) |
| 0.0 |
0.1 |
GO:0006002 |
fructose 6-phosphate metabolic process(GO:0006002) |
| 0.0 |
0.1 |
GO:0031581 |
hemidesmosome assembly(GO:0031581) |
| 0.0 |
0.0 |
GO:0010814 |
substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.0 |
0.1 |
GO:0048681 |
negative regulation of axon regeneration(GO:0048681) |
| 0.0 |
0.3 |
GO:0070207 |
protein homotrimerization(GO:0070207) |
| 0.0 |
0.1 |
GO:0006013 |
mannose metabolic process(GO:0006013) |
| 0.0 |
0.3 |
GO:0007214 |
gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 |
0.1 |
GO:0034587 |
piRNA metabolic process(GO:0034587) |
| 0.0 |
0.1 |
GO:0085020 |
protein K6-linked ubiquitination(GO:0085020) |
| 0.0 |
0.1 |
GO:0021764 |
amygdala development(GO:0021764) |
| 0.0 |
0.0 |
GO:0010993 |
regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 0.0 |
0.1 |
GO:0045719 |
negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 |
0.0 |
GO:0007066 |
female meiosis sister chromatid cohesion(GO:0007066) |
| 0.0 |
0.1 |
GO:0030242 |
pexophagy(GO:0030242) |
| 0.0 |
0.0 |
GO:0002184 |
cytoplasmic translational termination(GO:0002184) |
| 0.0 |
0.1 |
GO:1900194 |
negative regulation of oocyte maturation(GO:1900194) |
| 0.0 |
0.1 |
GO:0051574 |
positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 |
0.0 |
GO:0052428 |
modulation of molecular function in other organism(GO:0044359) negative regulation of molecular function in other organism(GO:0044362) negative regulation of molecular function in other organism involved in symbiotic interaction(GO:0052204) modulation of molecular function in other organism involved in symbiotic interaction(GO:0052205) negative regulation by host of symbiont molecular function(GO:0052405) modification by host of symbiont molecular function(GO:0052428) |
| 0.0 |
0.1 |
GO:0010820 |
positive regulation of T cell chemotaxis(GO:0010820) |
| 0.0 |
0.3 |
GO:0010829 |
negative regulation of glucose transport(GO:0010829) |
| 0.0 |
0.0 |
GO:0038128 |
ERBB2 signaling pathway(GO:0038128) |
| 0.0 |
0.1 |
GO:0003199 |
endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) endocardial cushion to mesenchymal transition(GO:0090500) positive regulation of blood vessel remodeling(GO:2000504) |