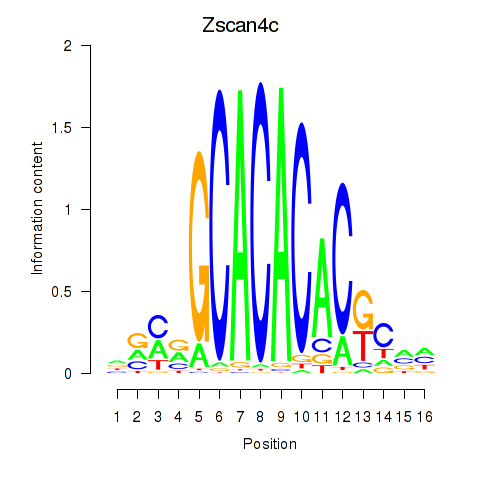
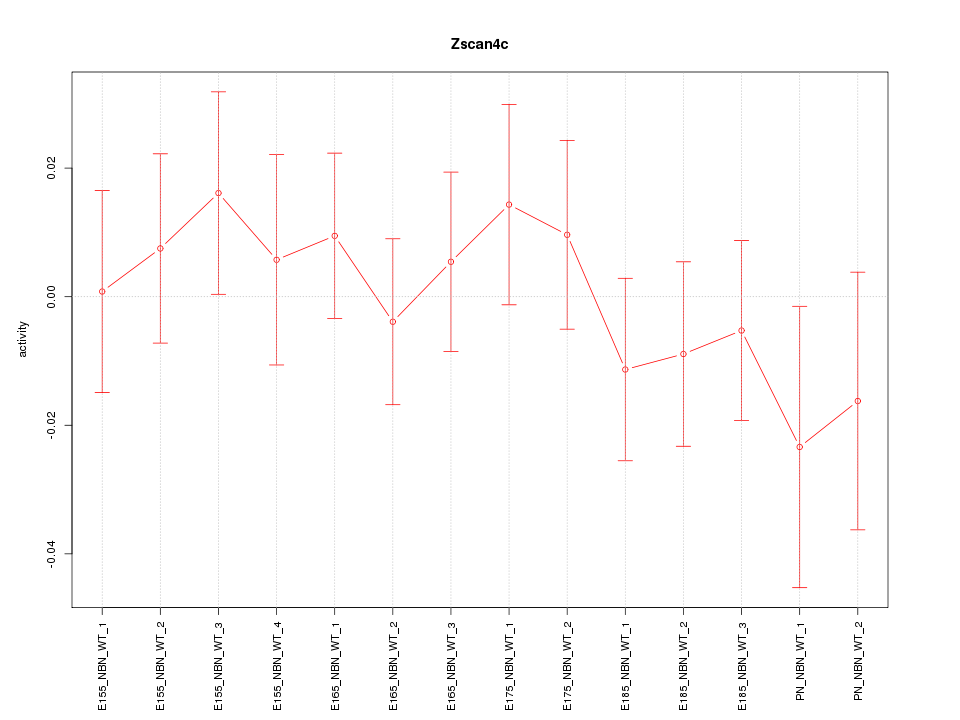
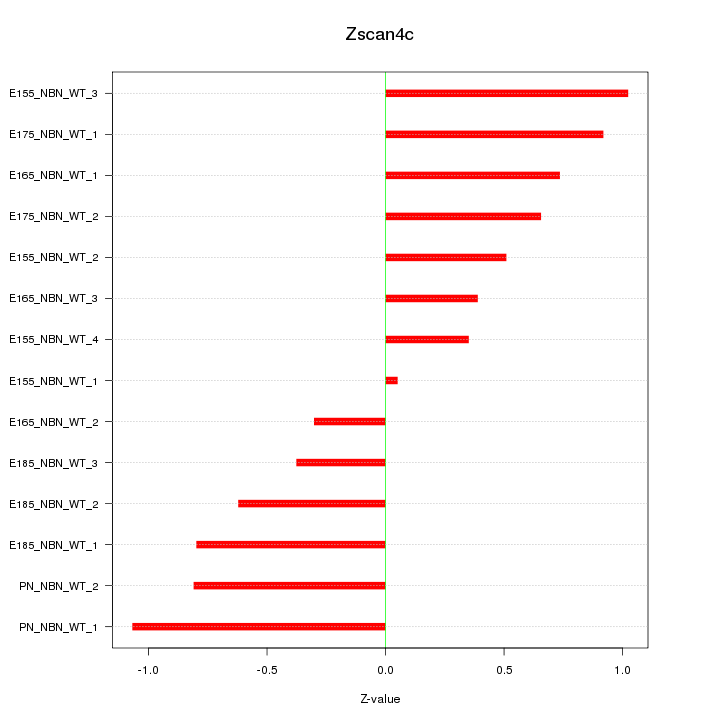
| 1.0 |
4.1 |
GO:0055095 |
lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) |
| 0.7 |
2.1 |
GO:0001928 |
regulation of exocyst assembly(GO:0001928) |
| 0.4 |
1.1 |
GO:0072070 |
establishment or maintenance of polarity of embryonic epithelium(GO:0016332) loop of Henle development(GO:0072070) metanephric loop of Henle development(GO:0072236) |
| 0.3 |
1.3 |
GO:0014846 |
esophagus smooth muscle contraction(GO:0014846) |
| 0.3 |
1.0 |
GO:2000501 |
natural killer cell chemotaxis(GO:0035747) regulation of natural killer cell chemotaxis(GO:2000501) |
| 0.3 |
2.0 |
GO:0048934 |
peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.3 |
1.0 |
GO:1901731 |
calcium-mediated signaling using extracellular calcium source(GO:0035585) positive regulation of platelet aggregation(GO:1901731) |
| 0.2 |
1.9 |
GO:0097264 |
self proteolysis(GO:0097264) |
| 0.2 |
1.5 |
GO:0061092 |
involuntary skeletal muscle contraction(GO:0003011) regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.2 |
1.0 |
GO:0089711 |
L-glutamate transmembrane transport(GO:0089711) |
| 0.2 |
3.4 |
GO:0050650 |
chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.2 |
0.2 |
GO:0070858 |
negative regulation of bile acid biosynthetic process(GO:0070858) negative regulation of bile acid metabolic process(GO:1904252) |
| 0.2 |
0.5 |
GO:1902277 |
negative regulation of pancreatic amylase secretion(GO:1902277) |
| 0.1 |
0.7 |
GO:0019659 |
lactate biosynthetic process from pyruvate(GO:0019244) lactate biosynthetic process(GO:0019249) glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.1 |
1.3 |
GO:0021869 |
forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.1 |
1.0 |
GO:0007379 |
segment specification(GO:0007379) |
| 0.1 |
5.3 |
GO:0046513 |
ceramide biosynthetic process(GO:0046513) |
| 0.1 |
0.4 |
GO:0061073 |
ciliary body morphogenesis(GO:0061073) |
| 0.1 |
0.5 |
GO:0007197 |
adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.1 |
0.2 |
GO:0046098 |
guanine metabolic process(GO:0046098) |
| 0.1 |
1.2 |
GO:0010669 |
epithelial structure maintenance(GO:0010669) |
| 0.1 |
0.9 |
GO:0035020 |
regulation of Rac protein signal transduction(GO:0035020) |
| 0.1 |
1.0 |
GO:0021542 |
dentate gyrus development(GO:0021542) |
| 0.0 |
2.9 |
GO:0010923 |
negative regulation of phosphatase activity(GO:0010923) |
| 0.0 |
0.1 |
GO:0006072 |
glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.0 |
0.2 |
GO:0019800 |
peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 |
1.1 |
GO:0007200 |
phospholipase C-activating G-protein coupled receptor signaling pathway(GO:0007200) |
| 0.0 |
0.1 |
GO:0071294 |
cellular response to zinc ion(GO:0071294) |
| 0.0 |
0.1 |
GO:1900116 |
sequestering of extracellular ligand from receptor(GO:0035581) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 |
0.7 |
GO:0090004 |
positive regulation of establishment of protein localization to plasma membrane(GO:0090004) positive regulation of potassium ion transmembrane transport(GO:1901381) |
| 0.0 |
0.4 |
GO:0010569 |
regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 |
0.1 |
GO:2001025 |
positive regulation of response to drug(GO:2001025) |
| 0.0 |
0.9 |
GO:0003073 |
regulation of systemic arterial blood pressure(GO:0003073) |
| 0.0 |
0.1 |
GO:0048170 |
positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |