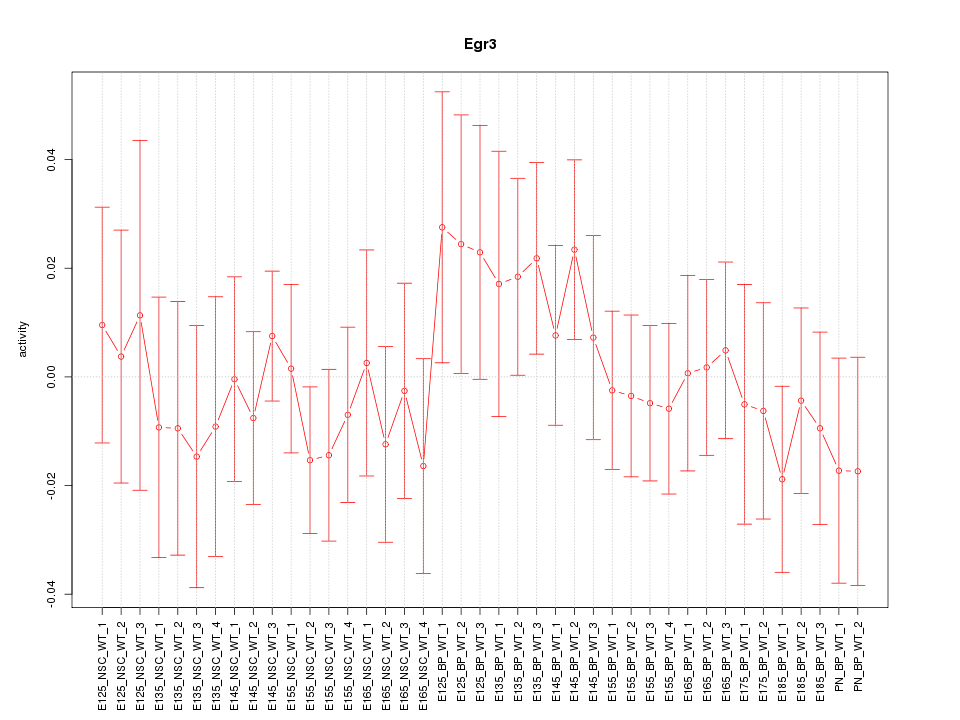
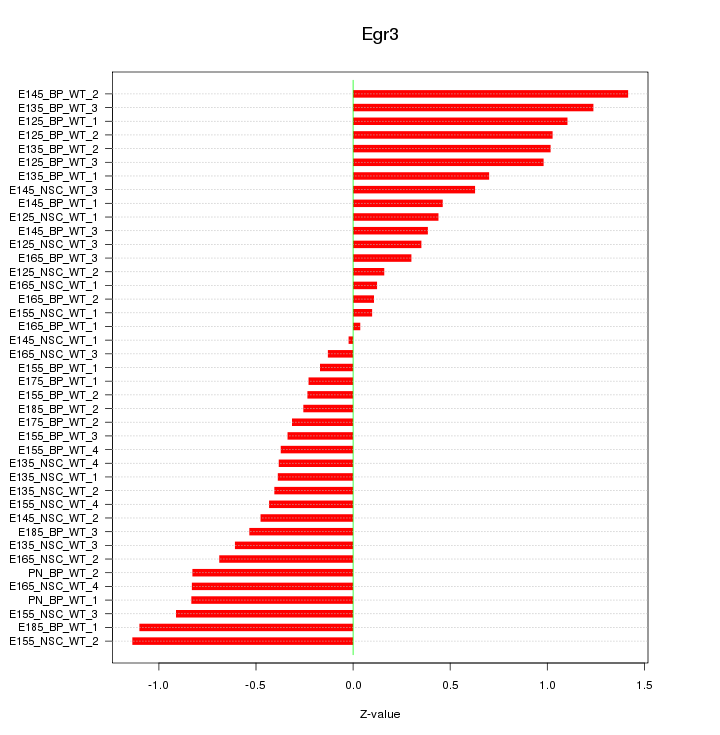
Motif ID: Egr3
Z-value: 0.653
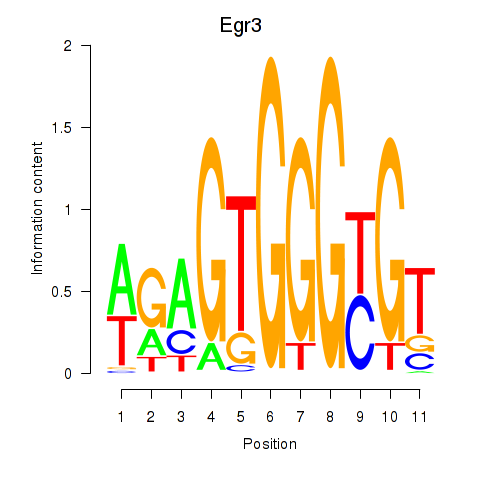
Transcription factors associated with Egr3:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Egr3 | ENSMUSG00000033730.3 | Egr3 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Egr3 | mm10_v2_chr14_+_70077375_70077445 | 0.29 | 6.4e-02 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.5 | 1.6 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.5 | 1.9 | GO:0003360 | brainstem development(GO:0003360) |
| 0.3 | 0.9 | GO:0060023 | soft palate development(GO:0060023) |
| 0.3 | 2.2 | GO:1990416 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.2 | 1.6 | GO:0000320 | re-entry into mitotic cell cycle(GO:0000320) |
| 0.2 | 2.0 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.2 | 1.1 | GO:2000325 | regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.2 | 1.1 | GO:0007403 | glial cell fate determination(GO:0007403) |
| 0.2 | 1.1 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.2 | 2.2 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.2 | 1.0 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.2 | 1.0 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.2 | 0.9 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.2 | 0.6 | GO:2000705 | regulation of dense core granule biogenesis(GO:2000705) |
| 0.2 | 0.5 | GO:1903334 | positive regulation of protein folding(GO:1903334) |
| 0.1 | 0.4 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.1 | 0.4 | GO:0010871 | negative regulation of receptor biosynthetic process(GO:0010871) |
| 0.1 | 0.3 | GO:2000812 | response to rapamycin(GO:1901355) regulation of barbed-end actin filament capping(GO:2000812) |
| 0.1 | 0.3 | GO:0045819 | plasmacytoid dendritic cell activation(GO:0002270) positive regulation of glycogen catabolic process(GO:0045819) |
| 0.1 | 0.8 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.1 | 0.5 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.1 | 0.4 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.1 | 1.8 | GO:0010763 | positive regulation of fibroblast migration(GO:0010763) |
| 0.1 | 0.3 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.1 | 0.3 | GO:0000189 | MAPK import into nucleus(GO:0000189) |
| 0.1 | 3.3 | GO:0007200 | phospholipase C-activating G-protein coupled receptor signaling pathway(GO:0007200) |
| 0.0 | 2.2 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.0 | 0.4 | GO:0097623 | potassium ion export across plasma membrane(GO:0097623) |
| 0.0 | 0.9 | GO:0001516 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.0 | 0.2 | GO:0060578 | extraocular skeletal muscle development(GO:0002074) subthalamic nucleus development(GO:0021763) left lung morphogenesis(GO:0060460) pulmonary vein morphogenesis(GO:0060577) superior vena cava morphogenesis(GO:0060578) |
| 0.0 | 0.3 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.0 | 2.3 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.8 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.9 | GO:0071364 | response to epidermal growth factor(GO:0070849) cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.0 | 0.3 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) negative regulation of autophagosome assembly(GO:1902902) |
| 0.0 | 0.5 | GO:0043486 | histone exchange(GO:0043486) |
| 0.0 | 3.0 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.7 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.1 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.1 | GO:0010757 | arachidonic acid metabolite production involved in inflammatory response(GO:0002538) negative regulation of plasminogen activation(GO:0010757) regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.0 | 0.3 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 0.4 | GO:0006378 | mRNA polyadenylation(GO:0006378) |
| 0.0 | 0.4 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.0 | 0.0 | GO:0045014 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) detection of glucose(GO:0051594) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
| 0.2 | 2.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.2 | 1.8 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.2 | 1.1 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.1 | 2.2 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.1 | 0.3 | GO:0031417 | NatC complex(GO:0031417) |
| 0.1 | 0.5 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.3 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.3 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.4 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 1.6 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.5 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.1 | GO:1990745 | EARP complex(GO:1990745) |
| 0.0 | 0.3 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.3 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.4 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.3 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.3 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.1 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.0 | 0.2 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 0.2 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.2 | GO:0032156 | septin cytoskeleton(GO:0032156) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.9 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.2 | 0.9 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.2 | 0.9 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.2 | 1.8 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.2 | 2.0 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.2 | 3.3 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.2 | 0.5 | GO:0019153 | protein-disulfide reductase (glutathione) activity(GO:0019153) |
| 0.1 | 4.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 0.5 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.1 | 0.3 | GO:0000402 | open form four-way junction DNA binding(GO:0000401) crossed form four-way junction DNA binding(GO:0000402) |
| 0.1 | 2.1 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 0.4 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.1 | 1.0 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 0.6 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.1 | 1.1 | GO:0001163 | RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.1 | 0.7 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.1 | 1.6 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 0.4 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.1 | 2.5 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 2.5 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 1.0 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 2.2 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.7 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.6 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.1 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.4 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.0 | 0.9 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 1.1 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.3 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.8 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.4 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.3 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.2 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.1 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 0.0 | 0.0 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.0 | 0.0 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.0 | 0.2 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.3 | GO:0034185 | apolipoprotein binding(GO:0034185) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | SA_REG_CASCADE_OF_CYCLIN_EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 3.9 | SIG_REGULATION_OF_THE_ACTIN_CYTOSKELETON_BY_RHO_GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 1.1 | PID_ERB_GENOMIC_PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 2.0 | NABA_PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.6 | PID_RXR_VDR_PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.8 | PID_NFKAPPAB_CANONICAL_PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 1.2 | PID_IFNG_PATHWAY | IFN-gamma pathway |
| 0.0 | 0.4 | PID_CIRCADIAN_PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.3 | PID_TOLL_ENDOGENOUS_PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.9 | PID_ILK_PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.3 | PID_SYNDECAN_2_PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 1.1 | PID_BETA_CATENIN_NUC_PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.8 | REACTOME_PRESYNAPTIC_NICOTINIC_ACETYLCHOLINE_RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 2.2 | REACTOME_INTERACTION_BETWEEN_L1_AND_ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 1.6 | REACTOME_PRE_NOTCH_TRANSCRIPTION_AND_TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.1 | 1.0 | REACTOME_GENERATION_OF_SECOND_MESSENGER_MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.6 | REACTOME_ADVANCED_GLYCOSYLATION_ENDPRODUCT_RECEPTOR_SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 1.6 | REACTOME_HS_GAG_BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.5 | REACTOME_CALNEXIN_CALRETICULIN_CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.8 | REACTOME_OXYGEN_DEPENDENT_PROLINE_HYDROXYLATION_OF_HYPOXIA_INDUCIBLE_FACTOR_ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 2.0 | REACTOME_G_ALPHA_Z_SIGNALLING_EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.3 | REACTOME_CHYLOMICRON_MEDIATED_LIPID_TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.4 | REACTOME_PROCESSING_OF_INTRONLESS_PRE_MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 2.5 | REACTOME_DIABETES_PATHWAYS | Genes involved in Diabetes pathways |
| 0.0 | 0.4 | REACTOME_ENDOSOMAL_SORTING_COMPLEX_REQUIRED_FOR_TRANSPORT_ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 1.0 | REACTOME_NUCLEAR_RECEPTOR_TRANSCRIPTION_PATHWAY | Genes involved in Nuclear Receptor transcription pathway |