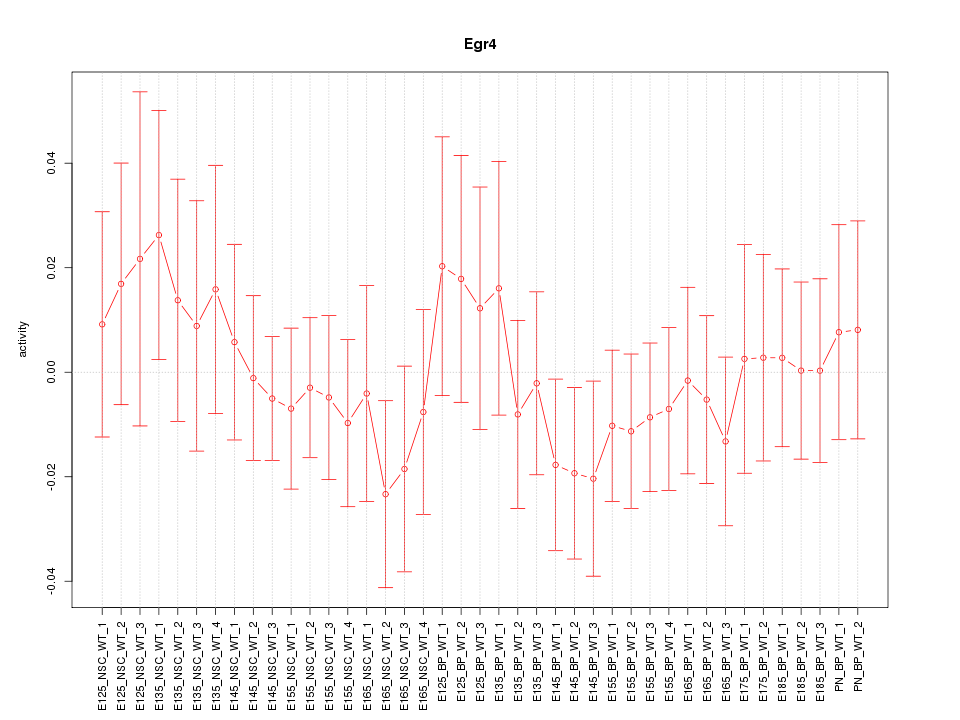
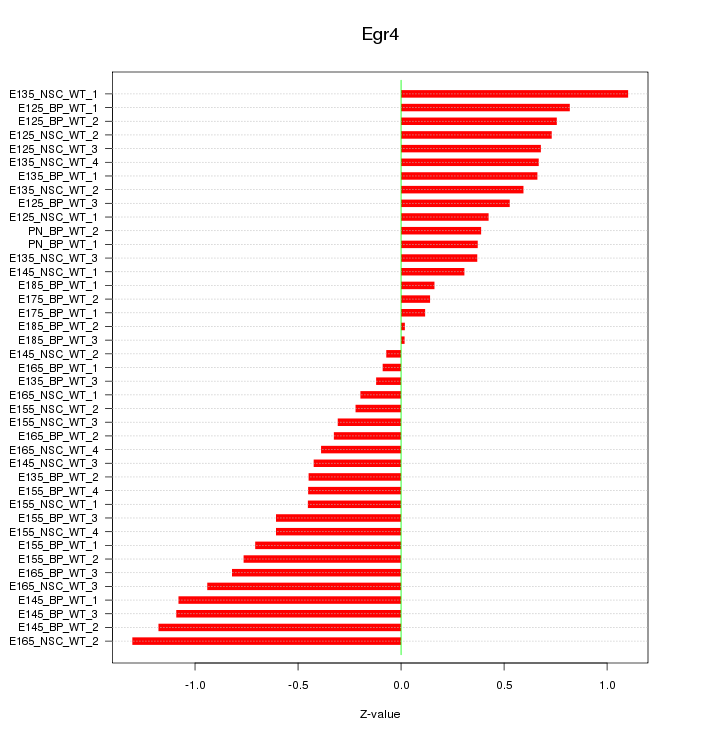
Motif ID: Egr4
Z-value: 0.621
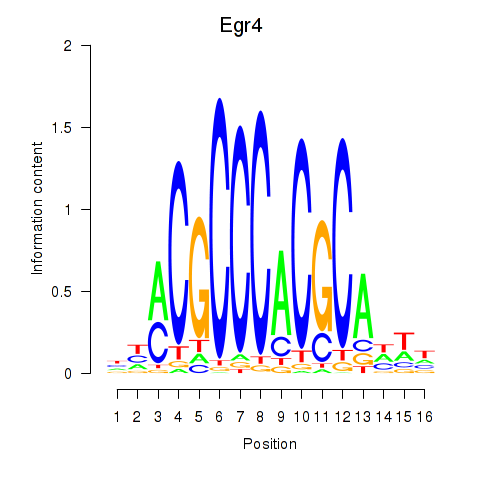
Transcription factors associated with Egr4:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Egr4 | ENSMUSG00000071341.3 | Egr4 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 5.2 | GO:2000325 | regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.8 | 3.1 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.4 | 1.2 | GO:0045819 | plasmacytoid dendritic cell activation(GO:0002270) positive regulation of glycogen catabolic process(GO:0045819) |
| 0.4 | 2.7 | GO:0060903 | positive regulation of meiosis I(GO:0060903) |
| 0.3 | 0.9 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.3 | 0.9 | GO:0003100 | angiotensin mediated vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001998) regulation of blood vessel size by renin-angiotensin(GO:0002034) renal control of peripheral vascular resistance involved in regulation of systemic arterial blood pressure(GO:0003072) regulation of systemic arterial blood pressure by endothelin(GO:0003100) beta selection(GO:0043366) response to chemokine(GO:1990868) cellular response to chemokine(GO:1990869) |
| 0.3 | 4.4 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.2 | 0.7 | GO:1904975 | response to bleomycin(GO:1904975) cellular response to bleomycin(GO:1904976) |
| 0.2 | 1.5 | GO:0000050 | urea cycle(GO:0000050) urea metabolic process(GO:0019627) |
| 0.2 | 0.7 | GO:0002572 | pro-T cell differentiation(GO:0002572) |
| 0.2 | 0.8 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.2 | 1.4 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.2 | 3.5 | GO:0060044 | negative regulation of cardiac muscle cell proliferation(GO:0060044) |
| 0.1 | 0.4 | GO:0045014 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) detection of glucose(GO:0051594) |
| 0.1 | 0.5 | GO:2000383 | regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.1 | 0.3 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.1 | 0.3 | GO:0010360 | negative regulation of anion channel activity(GO:0010360) |
| 0.1 | 1.8 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.1 | 1.1 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 0.3 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 0.1 | 1.6 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.1 | 0.6 | GO:1902510 | regulation of apoptotic DNA fragmentation(GO:1902510) |
| 0.1 | 0.2 | GO:0044340 | canonical Wnt signaling pathway involved in regulation of cell proliferation(GO:0044340) |
| 0.1 | 0.3 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.1 | 0.4 | GO:1900016 | negative regulation of cytokine production involved in inflammatory response(GO:1900016) |
| 0.0 | 0.5 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.0 | 0.8 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.6 | GO:0030238 | male sex determination(GO:0030238) |
| 0.0 | 1.4 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 1.3 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.6 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.2 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.0 | 1.9 | GO:0070613 | regulation of protein processing(GO:0070613) |
| 0.0 | 0.8 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.6 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) |
| 0.0 | 1.7 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) |
| 0.0 | 0.7 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
| 0.0 | 1.6 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.4 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.0 | 0.4 | GO:0001967 | suckling behavior(GO:0001967) |
| 0.0 | 0.8 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.6 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 | 0.8 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 | 0.1 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.0 | 0.5 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.7 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.0 | 0.0 | GO:0000189 | MAPK import into nucleus(GO:0000189) |
| 0.0 | 1.7 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.5 | 1.8 | GO:0002111 | BRCA2-BRAF35 complex(GO:0002111) |
| 0.3 | 0.8 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 1.3 | GO:0089701 | U2AF(GO:0089701) |
| 0.1 | 1.1 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.1 | 1.6 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.1 | 0.5 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.5 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.8 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.5 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.0 | 0.3 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 0.5 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.7 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 5.6 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.1 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.3 | GO:0098839 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.0 | 0.3 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 0.5 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 2.3 | GO:0001726 | ruffle(GO:0001726) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0000402 | open form four-way junction DNA binding(GO:0000401) crossed form four-way junction DNA binding(GO:0000402) |
| 0.2 | 4.4 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.2 | 0.6 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.2 | 5.2 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.1 | 0.6 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.1 | 1.3 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.1 | 0.9 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.1 | 1.8 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.1 | 1.5 | GO:0001013 | RNA polymerase I regulatory region DNA binding(GO:0001013) |
| 0.1 | 0.3 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.1 | 0.3 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 1.1 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.1 | GO:0005118 | sevenless binding(GO:0005118) |
| 0.0 | 0.8 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.5 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.2 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 2.0 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 3.9 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.8 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 1.3 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.7 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.4 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.0 | 1.5 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.3 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 3.1 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.1 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.8 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.5 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.7 | GO:0035064 | methylated histone binding(GO:0035064) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.1 | PID_TOLL_ENDOGENOUS_PATHWAY | Endogenous TLR signaling |
| 0.1 | 0.8 | ST_INTERFERON_GAMMA_PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.7 | PID_RET_PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 1.3 | PID_NFKAPPAB_CANONICAL_PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 4.4 | PID_AR_PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 5.2 | PID_BETA_CATENIN_NUC_PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.9 | PID_SMAD2_3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 1.9 | PID_HNF3B_PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.5 | PID_BETA_CATENIN_DEG_PATHWAY | Degradation of beta catenin |
| 0.0 | 0.7 | PID_CIRCADIAN_PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.6 | PID_CERAMIDE_PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.6 | PID_IL2_STAT5_PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.6 | PID_P75_NTR_PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 1.5 | PID_MYC_ACTIV_PATHWAY | Validated targets of C-MYC transcriptional activation |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.6 | REACTOME_DOWNREGULATION_OF_SMAD2_3_SMAD4_TRANSCRIPTIONAL_ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.1 | 1.3 | REACTOME_ADVANCED_GLYCOSYLATION_ENDPRODUCT_RECEPTOR_SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 0.8 | REACTOME_REGULATION_OF_IFNG_SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 0.5 | REACTOME_E2F_ENABLED_INHIBITION_OF_PRE_REPLICATION_COMPLEX_FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 0.9 | REACTOME_TGF_BETA_RECEPTOR_SIGNALING_IN_EMT_EPITHELIAL_TO_MESENCHYMAL_TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.4 | REACTOME_REGULATION_OF_GENE_EXPRESSION_IN_BETA_CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 1.3 | REACTOME_MRNA_3_END_PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.8 | REACTOME_DEADENYLATION_OF_MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.5 | REACTOME_KINESINS | Genes involved in Kinesins |
| 0.0 | 1.7 | REACTOME_FACTORS_INVOLVED_IN_MEGAKARYOCYTE_DEVELOPMENT_AND_PLATELET_PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.3 | REACTOME_CTNNB1_PHOSPHORYLATION_CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.3 | REACTOME_SIGNALING_BY_BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.2 | REACTOME_GLYCOGEN_BREAKDOWN_GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 1.0 | REACTOME_MHC_CLASS_II_ANTIGEN_PRESENTATION | Genes involved in MHC class II antigen presentation |