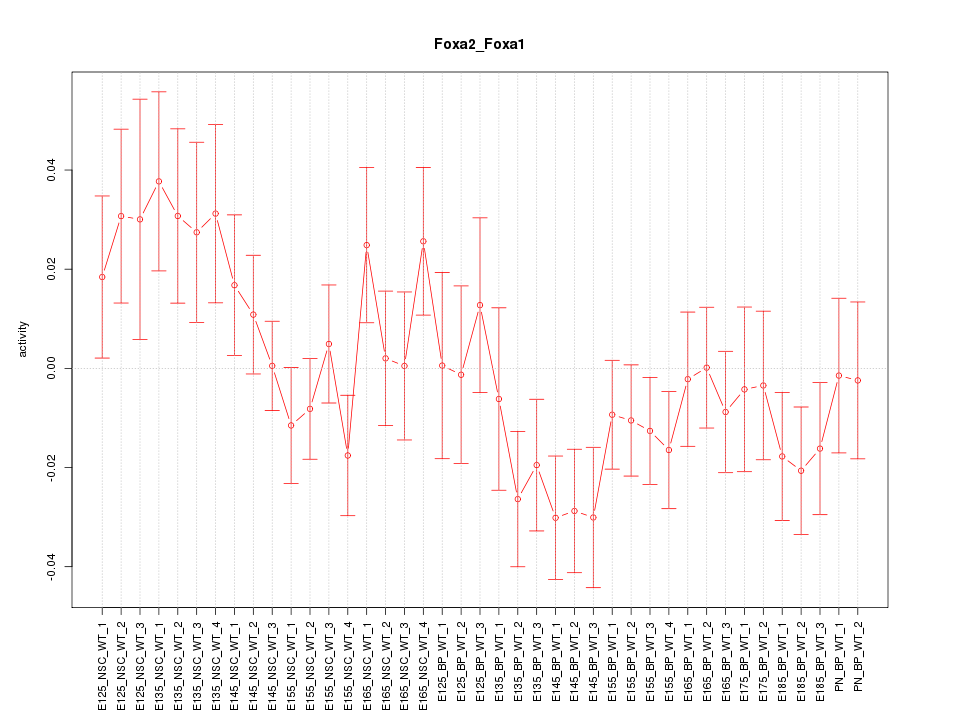
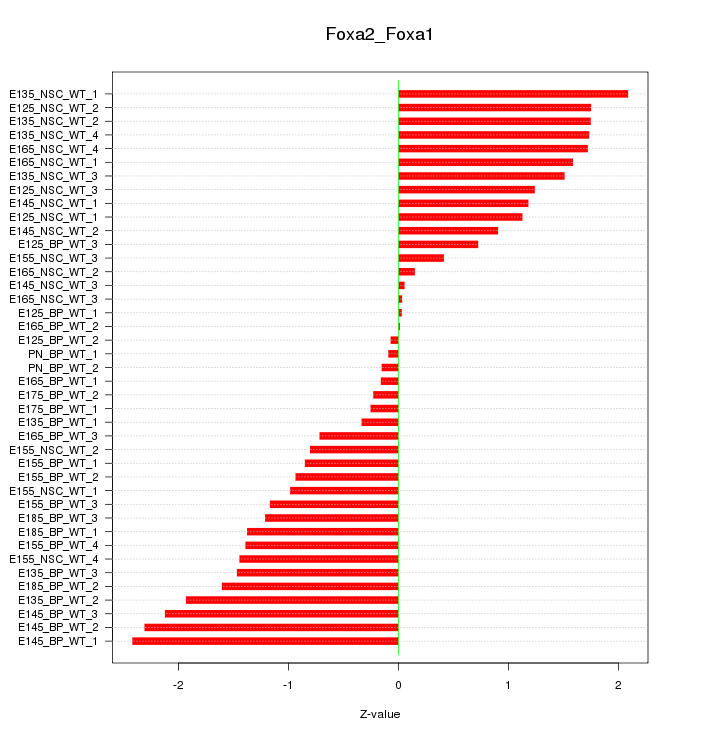
Motif ID: Foxa2_Foxa1
Z-value: 1.249
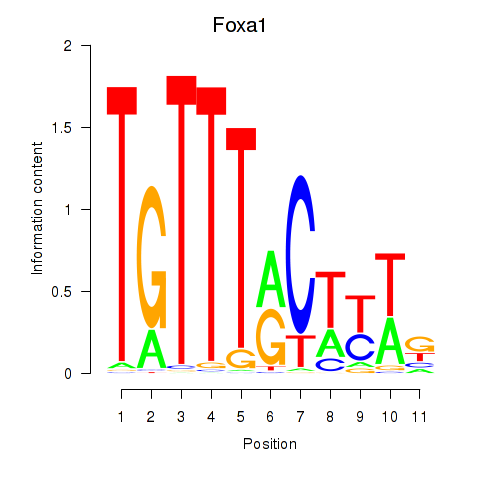
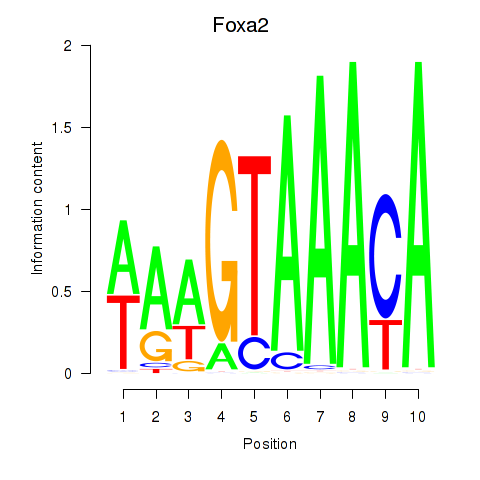
Transcription factors associated with Foxa2_Foxa1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Foxa1 | ENSMUSG00000035451.6 | Foxa1 |
| Foxa2 | ENSMUSG00000037025.5 | Foxa2 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Foxa2 | mm10_v2_chr2_-_148045891_148045948 | 0.28 | 7.2e-02 | Click! |
| Foxa1 | mm10_v2_chr12_-_57546121_57546141 | 0.26 | 1.0e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.7 | 11.1 | GO:0021759 | globus pallidus development(GO:0021759) |
| 3.4 | 13.8 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 2.0 | 6.1 | GO:1902524 | interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) positive regulation of protein K48-linked ubiquitination(GO:1902524) |
| 2.0 | 7.9 | GO:0046552 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 1.8 | 5.5 | GO:0035790 | platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) |
| 1.8 | 9.0 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 1.7 | 5.0 | GO:0072076 | hyaluranon cable assembly(GO:0036118) nephrogenic mesenchyme development(GO:0072076) negative regulation of glomerular mesangial cell proliferation(GO:0072125) negative regulation of glomerulus development(GO:0090194) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 1.5 | 4.4 | GO:0097402 | neuroblast migration(GO:0097402) |
| 1.5 | 16.0 | GO:0042473 | outer ear morphogenesis(GO:0042473) |
| 1.4 | 4.3 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 1.3 | 5.2 | GO:0003278 | apoptotic process involved in heart morphogenesis(GO:0003278) |
| 1.3 | 3.8 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 1.1 | 5.7 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 1.1 | 3.4 | GO:0014738 | regulation of muscle hyperplasia(GO:0014738) muscle hyperplasia(GO:0014900) |
| 1.0 | 2.9 | GO:0042320 | regulation of circadian sleep/wake cycle, REM sleep(GO:0042320) circadian sleep/wake cycle, REM sleep(GO:0042747) positive regulation of circadian sleep/wake cycle, sleep(GO:0045938) |
| 0.9 | 2.7 | GO:0060023 | soft palate development(GO:0060023) |
| 0.9 | 9.8 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.8 | 8.4 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.7 | 8.1 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.7 | 5.8 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.7 | 2.9 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.7 | 2.1 | GO:0045819 | plasmacytoid dendritic cell activation(GO:0002270) positive regulation of glycogen catabolic process(GO:0045819) |
| 0.7 | 11.2 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.7 | 7.6 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.6 | 3.1 | GO:0015817 | glutamine transport(GO:0006868) histidine transport(GO:0015817) cellular response to potassium ion starvation(GO:0051365) |
| 0.6 | 1.2 | GO:0001705 | ectoderm formation(GO:0001705) |
| 0.6 | 1.2 | GO:0098528 | regulation of branch elongation involved in ureteric bud branching(GO:0072095) skeletal muscle fiber differentiation(GO:0098528) |
| 0.6 | 2.4 | GO:2000182 | regulation of progesterone biosynthetic process(GO:2000182) |
| 0.5 | 5.4 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.5 | 8.8 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.5 | 1.9 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.5 | 1.4 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.4 | 3.5 | GO:0071493 | cellular response to UV-B(GO:0071493) |
| 0.4 | 1.2 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.4 | 4.3 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.4 | 2.2 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) establishment of Sertoli cell barrier(GO:0097368) |
| 0.4 | 3.5 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.3 | 3.2 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.3 | 3.5 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.3 | 1.8 | GO:0003383 | apical constriction(GO:0003383) |
| 0.3 | 1.5 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.3 | 1.5 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.3 | 1.7 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.3 | 8.5 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.3 | 0.8 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 0.3 | 0.8 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.3 | 0.8 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.3 | 1.0 | GO:0002572 | pro-T cell differentiation(GO:0002572) |
| 0.2 | 1.4 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.2 | 2.2 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.2 | 2.9 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.2 | 0.7 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) odontoblast differentiation(GO:0071895) regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) |
| 0.2 | 2.7 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.2 | 3.9 | GO:1904355 | positive regulation of telomere capping(GO:1904355) |
| 0.2 | 1.1 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.2 | 1.1 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.2 | 1.5 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.2 | 1.4 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.2 | 1.0 | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) |
| 0.2 | 0.8 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.2 | 0.7 | GO:0046909 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.2 | 0.8 | GO:0006447 | regulation of translational initiation by iron(GO:0006447) positive regulation of translational initiation by iron(GO:0045994) |
| 0.2 | 0.8 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.2 | 2.2 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.2 | 0.2 | GO:0060681 | branch elongation involved in ureteric bud branching(GO:0060681) |
| 0.1 | 0.9 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.1 | 1.2 | GO:0015074 | DNA integration(GO:0015074) |
| 0.1 | 1.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 2.0 | GO:0035646 | endosome to melanosome transport(GO:0035646) endosome to pigment granule transport(GO:0043485) pigment granule maturation(GO:0048757) |
| 0.1 | 0.5 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.1 | 0.4 | GO:2000334 | response to linoleic acid(GO:0070543) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.1 | 0.9 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.1 | 1.0 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.1 | 0.6 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.1 | 0.4 | GO:0010920 | negative regulation of inositol phosphate biosynthetic process(GO:0010920) |
| 0.1 | 2.3 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.1 | 1.7 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.1 | 0.2 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.1 | 0.8 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.1 | 1.8 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 0.2 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) alveolar secondary septum development(GO:0061144) |
| 0.1 | 0.6 | GO:0021559 | trigeminal nerve development(GO:0021559) |
| 0.1 | 0.9 | GO:0019369 | drug metabolic process(GO:0017144) arachidonic acid metabolic process(GO:0019369) |
| 0.1 | 0.4 | GO:0015791 | polyol transport(GO:0015791) |
| 0.0 | 0.5 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.3 | GO:0021830 | interneuron migration from the subpallium to the cortex(GO:0021830) |
| 0.0 | 0.6 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.0 | 2.8 | GO:0048663 | neuron fate commitment(GO:0048663) |
| 0.0 | 1.8 | GO:0030500 | regulation of bone mineralization(GO:0030500) |
| 0.0 | 0.2 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 0.8 | GO:2001240 | negative regulation of signal transduction in absence of ligand(GO:1901099) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 0.3 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 2.0 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 | 4.5 | GO:0030326 | embryonic limb morphogenesis(GO:0030326) embryonic appendage morphogenesis(GO:0035113) |
| 0.0 | 0.5 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.6 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.0 | 2.0 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 1.0 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 | 0.2 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.0 | 1.6 | GO:0050982 | detection of mechanical stimulus(GO:0050982) |
| 0.0 | 10.5 | GO:0042060 | wound healing(GO:0042060) |
| 0.0 | 0.2 | GO:1901072 | glucosamine-containing compound catabolic process(GO:1901072) |
| 0.0 | 1.3 | GO:0060538 | skeletal muscle organ development(GO:0060538) |
| 0.0 | 0.3 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.0 | 0.7 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.6 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.1 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 2.5 | GO:0007098 | centrosome cycle(GO:0007098) |
| 0.0 | 0.9 | GO:0002066 | columnar/cuboidal epithelial cell development(GO:0002066) |
| 0.0 | 0.3 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.0 | 0.2 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 1.2 | GO:0050868 | negative regulation of T cell activation(GO:0050868) |
| 0.0 | 0.1 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.0 | 0.2 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.6 | GO:0060706 | cell differentiation involved in embryonic placenta development(GO:0060706) |
| 0.0 | 0.1 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.3 | GO:0033194 | response to hydroperoxide(GO:0033194) |
| 0.0 | 0.8 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.3 | GO:0030282 | bone mineralization(GO:0030282) |
| 0.0 | 0.3 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 0.6 | GO:0000086 | G2/M transition of mitotic cell cycle(GO:0000086) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 8.5 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 2.0 | 9.8 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 1.2 | 3.5 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.6 | 5.2 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.5 | 2.0 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.5 | 15.8 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.4 | 4.3 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.4 | 1.1 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.4 | 1.9 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.4 | 5.4 | GO:0042555 | MCM complex(GO:0042555) |
| 0.3 | 0.7 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.2 | 1.5 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.2 | 1.8 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.1 | 1.0 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.1 | 5.8 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.1 | 0.8 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.1 | 13.8 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.1 | 1.0 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 0.6 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.1 | 0.6 | GO:1990357 | terminal web(GO:1990357) |
| 0.1 | 0.4 | GO:0001652 | granular component(GO:0001652) |
| 0.1 | 1.0 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.1 | 0.8 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.1 | 6.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 5.5 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 0.3 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.1 | 3.0 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 1.7 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 1.8 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 1.1 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 0.2 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 1.9 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.8 | GO:0098839 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.0 | 0.2 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.0 | 3.1 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 1.0 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 3.4 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.6 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 1.2 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 13.1 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 1.2 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 1.5 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 9.7 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 1.8 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.4 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 5.1 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 1.2 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 5.0 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 2.8 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.6 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.2 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.5 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.5 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 2.8 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.5 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.5 | GO:0005811 | lipid particle(GO:0005811) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 21.8 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 2.9 | 11.5 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 1.3 | 3.8 | GO:0016015 | morphogen activity(GO:0016015) |
| 1.2 | 3.5 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 1.1 | 5.5 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 1.0 | 3.1 | GO:0015182 | L-asparagine transmembrane transporter activity(GO:0015182) |
| 0.9 | 10.8 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.9 | 4.3 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.7 | 2.1 | GO:0000402 | open form four-way junction DNA binding(GO:0000401) crossed form four-way junction DNA binding(GO:0000402) |
| 0.7 | 3.4 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.6 | 2.9 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.5 | 5.0 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.5 | 10.6 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.4 | 1.7 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.4 | 1.1 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.4 | 1.8 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.3 | 0.9 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.3 | 1.9 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.3 | 8.5 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.2 | 6.1 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.2 | 5.0 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.2 | 4.9 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.2 | 4.4 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.2 | 2.1 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.2 | 6.9 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.2 | 1.1 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.2 | 3.5 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 0.8 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.1 | 0.8 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.1 | 1.0 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.1 | 2.4 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.1 | 0.4 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.1 | 0.3 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 1.7 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 1.1 | GO:0015197 | peptide transporter activity(GO:0015197) |
| 0.1 | 0.8 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.1 | 2.4 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 1.2 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.1 | 2.2 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 1.4 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 2.4 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.1 | 1.0 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 2.1 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 2.8 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.1 | 0.6 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 1.3 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.8 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.5 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 4.6 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.0 | 1.8 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 1.8 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 5.7 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 1.2 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.6 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.2 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.7 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.2 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.3 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.0 | 2.3 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.5 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.5 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.8 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.4 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.2 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 1.9 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.1 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 0.8 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 3.0 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 2.9 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.5 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.5 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.2 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 8.1 | GO:0000981 | RNA polymerase II transcription factor activity, sequence-specific DNA binding(GO:0000981) |
| 0.0 | 0.6 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 1.7 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 1.6 | GO:0000975 | regulatory region DNA binding(GO:0000975) transcription regulatory region DNA binding(GO:0044212) |
| 0.0 | 0.6 | GO:0016860 | intramolecular oxidoreductase activity(GO:0016860) |
| 0.0 | 0.1 | GO:0001846 | opsonin binding(GO:0001846) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 10.4 | SA_REG_CASCADE_OF_CYCLIN_EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.6 | 24.2 | PID_FOXM1_PATHWAY | FOXM1 transcription factor network |
| 0.5 | 3.5 | SA_G2_AND_M_PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.5 | 5.0 | PID_ALK2_PATHWAY | ALK2 signaling events |
| 0.4 | 5.2 | PID_INTEGRIN2_PATHWAY | Beta2 integrin cell surface interactions |
| 0.3 | 11.3 | PID_HNF3B_PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.2 | 5.2 | PID_PDGFRA_PATHWAY | PDGFR-alpha signaling pathway |
| 0.2 | 15.0 | PID_LYSOPHOSPHOLIPID_PATHWAY | LPA receptor mediated events |
| 0.2 | 5.8 | NABA_BASEMENT_MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 4.3 | PID_P38_MK2_PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 5.4 | PID_ATR_PATHWAY | ATR signaling pathway |
| 0.1 | 11.2 | PID_AR_PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 4.5 | ST_WNT_BETA_CATENIN_PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 2.4 | PID_HDAC_CLASSIII_PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 1.1 | PID_NFKAPPAB_ATYPICAL_PATHWAY | Atypical NF-kappaB pathway |
| 0.1 | 2.1 | PID_AMB2_NEUTROPHILS_PATHWAY | amb2 Integrin signaling |
| 0.1 | 6.9 | PID_BETA_CATENIN_NUC_PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 2.5 | PID_RET_PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 0.9 | PID_INSULIN_GLUCOSE_PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 1.7 | PID_IL2_PI3K_PATHWAY | IL2 signaling events mediated by PI3K |
| 0.1 | 0.3 | PID_ERBB_NETWORK_PATHWAY | ErbB receptor signaling network |
| 0.1 | 1.8 | SIG_INSULIN_RECEPTOR_PATHWAY_IN_CARDIAC_MYOCYTES | Genes related to the insulin receptor pathway |
| 0.1 | 1.7 | PID_ARF6_PATHWAY | Arf6 signaling events |
| 0.1 | 0.5 | PID_NECTIN_PATHWAY | Nectin adhesion pathway |
| 0.0 | 7.1 | NABA_ECM_REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.1 | PID_SYNDECAN_4_PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 1.4 | PID_ECADHERIN_NASCENT_AJ_PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.8 | PID_A6B1_A6B4_INTEGRIN_PATHWAY | a6b1 and a6b4 Integrin signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 8.1 | REACTOME_P2Y_RECEPTORS | Genes involved in P2Y receptors |
| 0.5 | 5.8 | REACTOME_ROLE_OF_SECOND_MESSENGERS_IN_NETRIN1_SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.4 | 10.4 | REACTOME_G0_AND_EARLY_G1 | Genes involved in G0 and Early G1 |
| 0.3 | 5.4 | REACTOME_UNWINDING_OF_DNA | Genes involved in Unwinding of DNA |
| 0.3 | 10.6 | REACTOME_DOWNREGULATION_OF_SMAD2_3_SMAD4_TRANSCRIPTIONAL_ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.2 | 3.4 | REACTOME_SIGNALING_BY_NODAL | Genes involved in Signaling by NODAL |
| 0.2 | 1.8 | REACTOME_GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.2 | 1.4 | REACTOME_APOPTOTIC_CLEAVAGE_OF_CELL_ADHESION_PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.2 | 1.9 | REACTOME_KERATAN_SULFATE_DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.2 | 5.8 | REACTOME_TIGHT_JUNCTION_INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 2.1 | REACTOME_APOPTOSIS_INDUCED_DNA_FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 6.9 | REACTOME_NCAM1_INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 1.7 | REACTOME_IL_7_SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 16.2 | REACTOME_CLASS_A1_RHODOPSIN_LIKE_RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |
| 0.1 | 1.7 | REACTOME_IRAK1_RECRUITS_IKK_COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.1 | 1.5 | REACTOME_CYTOCHROME_P450_ARRANGED_BY_SUBSTRATE_TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 2.9 | REACTOME_CIRCADIAN_REPRESSION_OF_EXPRESSION_BY_REV_ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 3.5 | REACTOME_G1_PHASE | Genes involved in G1 Phase |
| 0.1 | 4.5 | REACTOME_TRANSCRIPTIONAL_REGULATION_OF_WHITE_ADIPOCYTE_DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 3.3 | REACTOME_AMINO_ACID_TRANSPORT_ACROSS_THE_PLASMA_MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 1.8 | REACTOME_METAL_ION_SLC_TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 1.5 | REACTOME_KINESINS | Genes involved in Kinesins |
| 0.1 | 1.2 | REACTOME_REGULATION_OF_KIT_SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.1 | 0.4 | REACTOME_PASSIVE_TRANSPORT_BY_AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 1.3 | REACTOME_DIABETES_PATHWAYS | Genes involved in Diabetes pathways |
| 0.0 | 0.8 | REACTOME_MRNA_DECAY_BY_5_TO_3_EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 1.7 | REACTOME_PACKAGING_OF_TELOMERE_ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.8 | REACTOME_IONOTROPIC_ACTIVITY_OF_KAINATE_RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 10.5 | REACTOME_ANTIGEN_PROCESSING_UBIQUITINATION_PROTEASOME_DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 1.0 | REACTOME_DEADENYLATION_OF_MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 1.1 | REACTOME_GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.5 | REACTOME_NOTCH_HLH_TRANSCRIPTION_PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 1.1 | REACTOME_ACTIVATED_NOTCH1_TRANSMITS_SIGNAL_TO_THE_NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 2.3 | REACTOME_RESPONSE_TO_ELEVATED_PLATELET_CYTOSOLIC_CA2_ | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.6 | REACTOME_OTHER_SEMAPHORIN_INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.3 | REACTOME_SYNTHESIS_OF_PE | Genes involved in Synthesis of PE |
| 0.0 | 1.7 | REACTOME_FACTORS_INVOLVED_IN_MEGAKARYOCYTE_DEVELOPMENT_AND_PLATELET_PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.1 | REACTOME_CREATION_OF_C4_AND_C2_ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.3 | REACTOME_GRB2_EVENTS_IN_ERBB2_SIGNALING | Genes involved in GRB2 events in ERBB2 signaling |
| 0.0 | 0.5 | REACTOME_ADHERENS_JUNCTIONS_INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.2 | REACTOME_SIGNALING_BY_ACTIVATED_POINT_MUTANTS_OF_FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.2 | REACTOME_SYNTHESIS_OF_PC | Genes involved in Synthesis of PC |
| 0.0 | 0.5 | REACTOME_ION_TRANSPORT_BY_P_TYPE_ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 1.0 | REACTOME_MITOTIC_PROMETAPHASE | Genes involved in Mitotic Prometaphase |