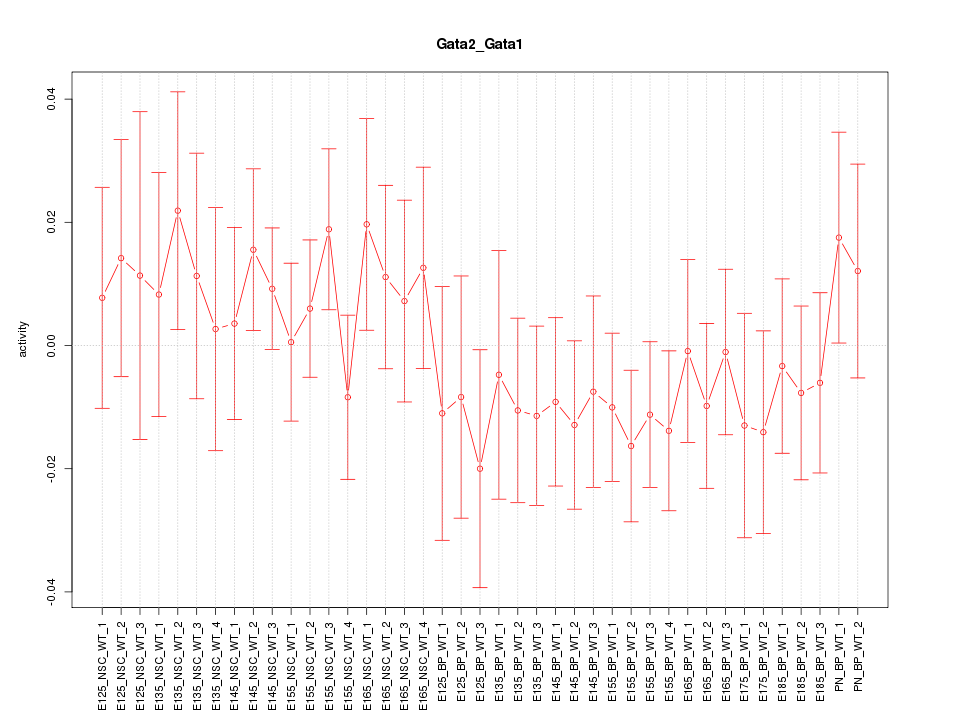
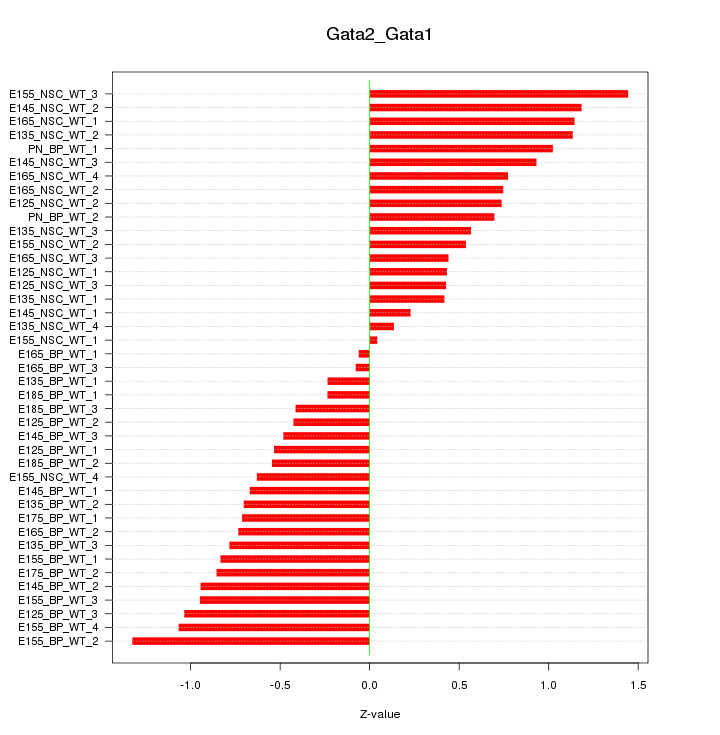
Motif ID: Gata2_Gata1
Z-value: 0.750
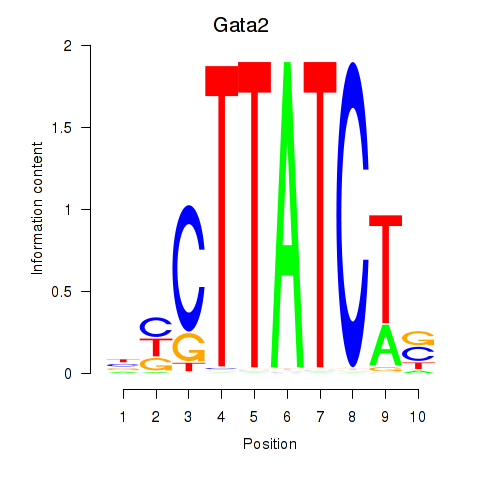
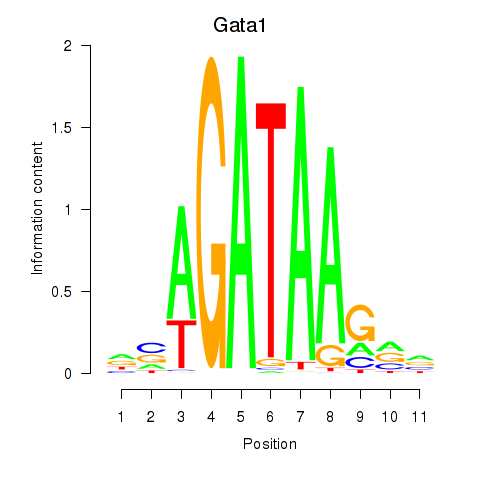
Transcription factors associated with Gata2_Gata1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Gata1 | ENSMUSG00000031162.8 | Gata1 |
| Gata2 | ENSMUSG00000015053.8 | Gata2 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Gata2 | mm10_v2_chr6_+_88198656_88198675 | 0.31 | 4.6e-02 | Click! |
| Gata1 | mm10_v2_chrX_-_7967817_7967910 | 0.22 | 1.7e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 6.2 | GO:0032650 | regulation of interleukin-1 alpha production(GO:0032650) positive regulation of interleukin-1 alpha production(GO:0032730) cardiac cell fate determination(GO:0060913) |
| 0.9 | 5.3 | GO:0001980 | regulation of systemic arterial blood pressure by ischemic conditions(GO:0001980) |
| 0.8 | 2.4 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.7 | 2.0 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.7 | 3.3 | GO:1903609 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 0.7 | 2.6 | GO:0051933 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 0.6 | 3.8 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.6 | 4.1 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.6 | 1.7 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.5 | 4.4 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.5 | 3.0 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.5 | 3.4 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.4 | 2.6 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.4 | 2.2 | GO:0002317 | plasma cell differentiation(GO:0002317) negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.3 | 2.4 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.3 | 1.0 | GO:0030300 | regulation of intestinal cholesterol absorption(GO:0030300) negative regulation of intestinal absorption(GO:1904479) |
| 0.2 | 3.0 | GO:0070995 | toxin metabolic process(GO:0009404) NADPH oxidation(GO:0070995) |
| 0.2 | 1.5 | GO:0097646 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.2 | 2.4 | GO:0070574 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.2 | 1.1 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) |
| 0.2 | 0.4 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.2 | 1.0 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.2 | 0.8 | GO:0010593 | negative regulation of lamellipodium assembly(GO:0010593) |
| 0.1 | 0.8 | GO:0033632 | regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.1 | 0.5 | GO:0097168 | condensed mesenchymal cell proliferation(GO:0072137) mesenchymal stem cell proliferation(GO:0097168) |
| 0.1 | 0.4 | GO:0035811 | negative regulation of urine volume(GO:0035811) |
| 0.1 | 0.3 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) |
| 0.1 | 0.3 | GO:0003057 | regulation of the force of heart contraction by chemical signal(GO:0003057) |
| 0.1 | 1.3 | GO:0018065 | protein-cofactor linkage(GO:0018065) |
| 0.1 | 1.4 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.1 | 0.8 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.1 | 1.8 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.1 | 0.3 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.1 | 1.3 | GO:0034377 | plasma lipoprotein particle assembly(GO:0034377) |
| 0.1 | 10.8 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.1 | 0.8 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 0.1 | 1.8 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.1 | 0.4 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.1 | 0.9 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.1 | 0.4 | GO:0000019 | regulation of mitotic recombination(GO:0000019) telomeric 3' overhang formation(GO:0031860) |
| 0.1 | 0.3 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.1 | 1.0 | GO:0046697 | decidualization(GO:0046697) |
| 0.1 | 0.4 | GO:0032261 | purine nucleotide salvage(GO:0032261) IMP salvage(GO:0032264) |
| 0.1 | 0.2 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.1 | 0.4 | GO:0006586 | tryptophan metabolic process(GO:0006568) indolalkylamine metabolic process(GO:0006586) |
| 0.0 | 0.4 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.0 | 1.0 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.0 | 0.4 | GO:0086023 | adrenergic receptor signaling pathway involved in heart process(GO:0086023) |
| 0.0 | 3.1 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.0 | 0.5 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 0.3 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 0.2 | GO:0046880 | regulation of gonadotropin secretion(GO:0032276) regulation of follicle-stimulating hormone secretion(GO:0046880) follicle-stimulating hormone secretion(GO:0046884) |
| 0.0 | 0.1 | GO:1904139 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.0 | 1.0 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 0.3 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.4 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.0 | 0.2 | GO:0015862 | uridine transport(GO:0015862) |
| 0.0 | 0.3 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 1.2 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 0.2 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.0 | 1.5 | GO:0021766 | hippocampus development(GO:0021766) |
| 0.0 | 0.1 | GO:0044539 | long-chain fatty acid import(GO:0044539) |
| 0.0 | 0.1 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) |
| 0.0 | 0.5 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.0 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.5 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.0 | 0.2 | GO:0031272 | regulation of pseudopodium assembly(GO:0031272) |
| 0.0 | 0.3 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 0.4 | GO:0030168 | platelet activation(GO:0030168) |
| 0.0 | 0.4 | GO:0021799 | cerebral cortex radially oriented cell migration(GO:0021799) centrosome localization(GO:0051642) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 5.3 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.7 | 10.6 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.5 | 3.2 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.3 | 3.3 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.2 | 2.4 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.1 | 2.4 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.1 | 1.0 | GO:0098533 | ATPase dependent transmembrane transport complex(GO:0098533) |
| 0.1 | 2.1 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 0.4 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.1 | 0.2 | GO:0043512 | inhibin-betaglycan-ActRII complex(GO:0034673) inhibin A complex(GO:0043512) |
| 0.1 | 0.3 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.1 | 0.3 | GO:0005587 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.1 | 1.1 | GO:0008305 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.1 | 0.4 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.5 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.2 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.2 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.0 | 4.4 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 1.0 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.6 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 3.4 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 1.6 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.2 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 2.0 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.0 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.3 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.2 | GO:0005839 | proteasome core complex(GO:0005839) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.3 | GO:0030172 | troponin C binding(GO:0030172) |
| 1.1 | 1.1 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.9 | 2.6 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.7 | 3.3 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.6 | 3.0 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.6 | 1.7 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.5 | 1.4 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.4 | 11.6 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.4 | 2.0 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.4 | 4.1 | GO:0019531 | secondary active sulfate transmembrane transporter activity(GO:0008271) oxalate transmembrane transporter activity(GO:0019531) |
| 0.3 | 2.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.3 | 1.7 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.3 | 3.4 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.3 | 1.0 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.2 | 0.9 | GO:0004921 | interleukin-11 receptor activity(GO:0004921) interleukin-11 binding(GO:0019970) |
| 0.2 | 1.3 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.2 | 6.2 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.2 | 1.0 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.1 | 0.4 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.1 | 0.4 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.1 | 1.9 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.3 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.1 | 2.4 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 1.1 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 0.3 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 2.4 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 0.3 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.1 | 0.8 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.1 | 0.2 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.1 | 0.4 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.1 | 0.4 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.1 | 0.4 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 0.4 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 0.1 | GO:0043758 | acetate-CoA ligase (ADP-forming) activity(GO:0043758) |
| 0.0 | 0.4 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.0 | 2.2 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 1.7 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 1.9 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.0 | 0.4 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.8 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.2 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 0.6 | GO:0005272 | sodium channel activity(GO:0005272) |
| 0.0 | 1.8 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.0 | 0.4 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.1 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.0 | 0.2 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 0.5 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.0 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 0.1 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.0 | 0.4 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.0 | 1.0 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 13.3 | GO:0043565 | sequence-specific DNA binding(GO:0043565) |
| 0.0 | 0.7 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.3 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.1 | GO:0070700 | BMP receptor binding(GO:0070700) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.3 | PID_WNT_CANONICAL_PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 7.0 | PID_MYC_ACTIV_PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 1.1 | PID_INTEGRIN_CS_PATHWAY | Integrin family cell surface interactions |
| 0.1 | 1.8 | NABA_PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.0 | PID_P38_MK2_PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 1.2 | PID_EPO_PATHWAY | EPO signaling pathway |
| 0.0 | 0.2 | PID_ALK2_PATHWAY | ALK2 signaling events |
| 0.0 | 5.1 | NABA_SECRETED_FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.4 | PID_LPA4_PATHWAY | LPA4-mediated signaling events |
| 0.0 | 1.2 | PID_TAP63_PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 1.8 | PID_BETA_CATENIN_NUC_PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.2 | PID_INTEGRIN2_PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 2.2 | NABA_ECM_AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.3 | PID_FRA_PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.4 | ST_MYOCYTE_AD_PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.3 | ST_WNT_BETA_CATENIN_PATHWAY | Wnt/beta-catenin Pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 6.2 | REACTOME_SYNTHESIS_SECRETION_AND_INACTIVATION_OF_GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.2 | 4.3 | REACTOME_STRIATED_MUSCLE_CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.2 | 7.5 | REACTOME_LIPID_DIGESTION_MOBILIZATION_AND_TRANSPORT | Genes involved in Lipid digestion, mobilization, and transport |
| 0.1 | 2.4 | REACTOME_ZINC_TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 1.9 | REACTOME_CS_DS_DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 3.0 | REACTOME_PHASE1_FUNCTIONALIZATION_OF_COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.1 | 1.5 | REACTOME_P130CAS_LINKAGE_TO_MAPK_SIGNALING_FOR_INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.1 | 2.0 | REACTOME_AMINE_COMPOUND_SLC_TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.1 | 0.2 | REACTOME_TRANSLOCATION_OF_ZAP_70_TO_IMMUNOLOGICAL_SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.1 | 2.6 | REACTOME_INHIBITION_OF_VOLTAGE_GATED_CA2_CHANNELS_VIA_GBETA_GAMMA_SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 1.4 | REACTOME_METABOLISM_OF_PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 1.2 | REACTOME_REGULATION_OF_KIT_SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.1 | 0.2 | REACTOME_GLYCOPROTEIN_HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 1.2 | REACTOME_ELONGATION_ARREST_AND_RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 3.8 | REACTOME_TRANSPORT_OF_INORGANIC_CATIONS_ANIONS_AND_AMINO_ACIDS_OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.4 | REACTOME_HOMOLOGOUS_RECOMBINATION_REPAIR_OF_REPLICATION_INDEPENDENT_DOUBLE_STRAND_BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.4 | REACTOME_AMINE_DERIVED_HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.1 | REACTOME_ADP_SIGNALLING_THROUGH_P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.0 | 0.5 | REACTOME_POST_CHAPERONIN_TUBULIN_FOLDING_PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.2 | REACTOME_INTRINSIC_PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.8 | REACTOME_OTHER_SEMAPHORIN_INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.4 | REACTOME_ADENYLATE_CYCLASE_ACTIVATING_PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.4 | REACTOME_PURINE_SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.3 | REACTOME_SYNTHESIS_OF_PE | Genes involved in Synthesis of PE |
| 0.0 | 1.0 | REACTOME_TRNA_AMINOACYLATION | Genes involved in tRNA Aminoacylation |
| 0.0 | 1.5 | REACTOME_G_ALPHA_S_SIGNALLING_EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.3 | REACTOME_NITRIC_OXIDE_STIMULATES_GUANYLATE_CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.0 | 0.4 | REACTOME_MYOGENESIS | Genes involved in Myogenesis |