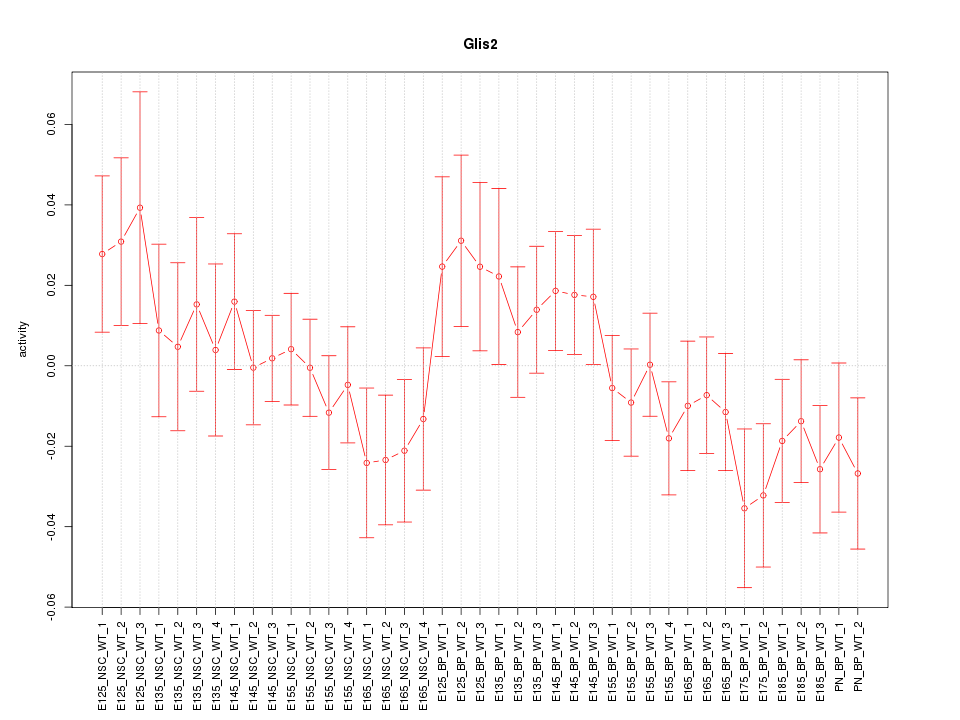
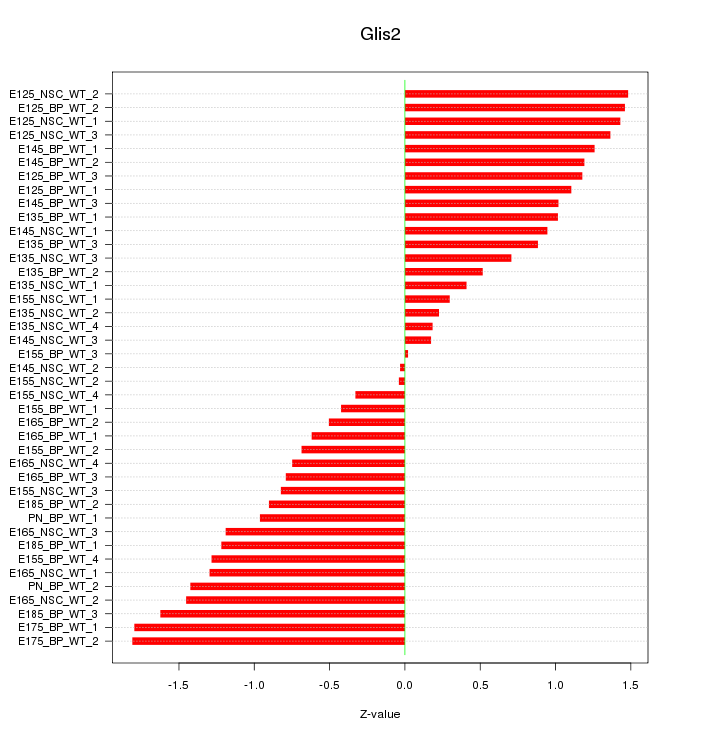
Motif ID: Glis2
Z-value: 1.027

Transcription factors associated with Glis2:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Glis2 | ENSMUSG00000014303.7 | Glis2 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Glis2 | mm10_v2_chr16_+_4594683_4594735 | -0.31 | 4.8e-02 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 18.1 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 1.8 | 7.2 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 1.6 | 4.9 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 1.3 | 5.2 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 1.2 | 15.2 | GO:0042573 | retinoic acid metabolic process(GO:0042573) |
| 1.0 | 2.9 | GO:2000501 | natural killer cell chemotaxis(GO:0035747) regulation of natural killer cell chemotaxis(GO:2000501) |
| 0.9 | 4.5 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.9 | 2.6 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 0.8 | 3.8 | GO:1905247 | positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 0.7 | 2.2 | GO:0043031 | regulation of phosphatidylcholine catabolic process(GO:0010899) negative regulation of macrophage activation(GO:0043031) receptor-mediated endocytosis of low-density lipoprotein particle involved in cholesterol transport(GO:0090118) regulation of microglial cell activation(GO:1903978) |
| 0.7 | 3.5 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.7 | 2.1 | GO:0010752 | negative regulation of antigen processing and presentation(GO:0002578) negative regulation of nitric oxide mediated signal transduction(GO:0010751) regulation of cGMP-mediated signaling(GO:0010752) |
| 0.6 | 3.6 | GO:0032796 | uropod organization(GO:0032796) |
| 0.6 | 1.7 | GO:0001978 | regulation of systemic arterial blood pressure by carotid sinus baroreceptor feedback(GO:0001978) |
| 0.5 | 2.1 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.5 | 2.6 | GO:0033499 | galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.5 | 1.4 | GO:0060129 | thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) |
| 0.5 | 4.6 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.4 | 1.3 | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.4 | 2.6 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.4 | 2.1 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.4 | 1.5 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.4 | 1.9 | GO:0035984 | response to trichostatin A(GO:0035983) cellular response to trichostatin A(GO:0035984) |
| 0.4 | 3.6 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.4 | 1.4 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.3 | 2.2 | GO:0072086 | specification of loop of Henle identity(GO:0072086) |
| 0.3 | 0.9 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.3 | 1.4 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.3 | 1.2 | GO:0046898 | response to cycloheximide(GO:0046898) |
| 0.3 | 3.6 | GO:0090394 | negative regulation of excitatory postsynaptic potential(GO:0090394) |
| 0.3 | 1.0 | GO:0046552 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) retinal rod cell differentiation(GO:0060221) |
| 0.2 | 3.7 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.2 | 3.2 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.2 | 1.9 | GO:0015074 | DNA integration(GO:0015074) |
| 0.2 | 2.5 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.2 | 7.9 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.2 | 2.0 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.2 | 1.1 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.2 | 5.1 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.2 | 0.6 | GO:0021577 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.2 | 0.6 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.2 | 1.7 | GO:0007512 | adult heart development(GO:0007512) |
| 0.2 | 1.3 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.2 | 2.8 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.2 | 1.3 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.2 | 0.5 | GO:1990859 | cellular response to endothelin(GO:1990859) |
| 0.2 | 2.0 | GO:0033008 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.1 | 0.6 | GO:0072369 | regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 0.1 | 0.4 | GO:1902022 | L-lysine transport(GO:1902022) |
| 0.1 | 1.0 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 1.8 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.1 | 1.5 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.1 | 0.8 | GO:0006621 | protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.1 | 3.0 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.1 | 0.9 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.1 | 0.9 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.1 | 1.0 | GO:0001771 | immunological synapse formation(GO:0001771) |
| 0.1 | 0.3 | GO:0044778 | meiotic DNA integrity checkpoint(GO:0044778) |
| 0.1 | 1.0 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.1 | 0.3 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.1 | 0.8 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.1 | 0.6 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 2.3 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.1 | 0.2 | GO:0042921 | glucocorticoid receptor signaling pathway(GO:0042921) |
| 0.1 | 0.2 | GO:0061181 | regulation of chondrocyte development(GO:0061181) |
| 0.1 | 2.2 | GO:0001964 | startle response(GO:0001964) |
| 0.1 | 2.8 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.1 | 1.6 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.2 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.0 | 0.1 | GO:0070844 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.0 | 1.0 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.3 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.0 | 2.2 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.4 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.5 | GO:2000821 | regulation of grooming behavior(GO:2000821) |
| 0.0 | 0.7 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.1 | GO:1902037 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 0.0 | 1.2 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
| 0.0 | 5.1 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.9 | GO:0043303 | mast cell activation involved in immune response(GO:0002279) mast cell degranulation(GO:0043303) |
| 0.0 | 1.3 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.1 | GO:2001185 | regulation of CD8-positive, alpha-beta T cell activation(GO:2001185) |
| 0.0 | 0.3 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.0 | 0.3 | GO:0042297 | vocal learning(GO:0042297) imitative learning(GO:0098596) |
| 0.0 | 0.1 | GO:0035547 | interferon-beta secretion(GO:0035546) regulation of interferon-beta secretion(GO:0035547) positive regulation of interferon-beta secretion(GO:0035549) CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 1.8 | GO:0050731 | positive regulation of peptidyl-tyrosine phosphorylation(GO:0050731) |
| 0.0 | 0.4 | GO:2000398 | regulation of T cell differentiation in thymus(GO:0033081) regulation of thymocyte aggregation(GO:2000398) |
| 0.0 | 1.2 | GO:0008306 | associative learning(GO:0008306) |
| 0.0 | 0.2 | GO:0032402 | melanosome transport(GO:0032402) |
| 0.0 | 0.9 | GO:0001885 | endothelial cell development(GO:0001885) |
| 0.0 | 1.5 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.7 | 2.0 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.6 | 4.5 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.6 | 7.9 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.5 | 2.0 | GO:0090537 | CERF complex(GO:0090537) |
| 0.4 | 3.4 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.4 | 2.2 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.3 | 4.6 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.3 | 3.6 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.3 | 0.9 | GO:0033193 | Lsd1/2 complex(GO:0033193) |
| 0.3 | 1.4 | GO:0072487 | MSL complex(GO:0072487) |
| 0.2 | 1.2 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.2 | 1.5 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.2 | 2.3 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.2 | 0.9 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.2 | 1.4 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.2 | 7.4 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.2 | 0.5 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.1 | 1.5 | GO:0034709 | methylosome(GO:0034709) |
| 0.1 | 1.8 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.1 | 1.7 | GO:0005675 | holo TFIIH complex(GO:0005675) |
| 0.1 | 1.3 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.1 | 1.2 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 0.8 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 1.0 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 5.1 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 2.2 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 2.3 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.1 | 3.0 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 1.0 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 5.2 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.6 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.3 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 0.3 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.8 | GO:0044306 | neuron projection terminus(GO:0044306) |
| 0.0 | 0.5 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 1.3 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.8 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 5.7 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 2.6 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 0.4 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 7.1 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.8 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 4.1 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.2 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 1.2 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.4 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 1.4 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 1.5 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.3 | GO:0016592 | mediator complex(GO:0016592) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 15.2 | GO:0019841 | retinol binding(GO:0019841) |
| 1.6 | 4.9 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 1.1 | 4.5 | GO:0031721 | hemoglobin alpha binding(GO:0031721) |
| 0.8 | 4.6 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.7 | 5.2 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) lysophosphatidic acid receptor activity(GO:0070915) |
| 0.7 | 2.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.7 | 2.6 | GO:1902379 | chemoattractant activity involved in axon guidance(GO:1902379) |
| 0.6 | 3.5 | GO:0051429 | corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.6 | 5.1 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.5 | 2.6 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.5 | 1.5 | GO:0035605 | peptidyl-cysteine S-nitrosylase activity(GO:0035605) |
| 0.5 | 2.0 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.5 | 3.4 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.5 | 1.4 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.4 | 2.2 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.4 | 1.3 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.4 | 2.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.4 | 1.7 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.3 | 3.6 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.3 | 0.9 | GO:0071936 | coreceptor activity involved in Wnt signaling pathway(GO:0071936) |
| 0.3 | 1.4 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.2 | 7.4 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.2 | 1.3 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.2 | 0.8 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.2 | 1.5 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.2 | 0.9 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.2 | 2.6 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.1 | 2.8 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 1.2 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.1 | 1.5 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.1 | 3.8 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 0.6 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.1 | 2.9 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.1 | 2.5 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 0.4 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.1 | 1.4 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.1 | 3.0 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 3.3 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 0.5 | GO:0031811 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) neuroligin family protein binding(GO:0097109) |
| 0.1 | 1.5 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 0.4 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 0.8 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.2 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 1.5 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.5 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.1 | GO:0033680 | ATP-dependent DNA/RNA helicase activity(GO:0033680) |
| 0.0 | 0.9 | GO:0042165 | neurotransmitter binding(GO:0042165) |
| 0.0 | 0.1 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.0 | 1.4 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 1.7 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 1.4 | GO:0001067 | regulatory region nucleic acid binding(GO:0001067) |
| 0.0 | 2.1 | GO:0000981 | RNA polymerase II transcription factor activity, sequence-specific DNA binding(GO:0000981) |
| 0.0 | 1.2 | GO:0020037 | heme binding(GO:0020037) |
| 0.0 | 5.3 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.4 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 1.3 | GO:0072341 | modified amino acid binding(GO:0072341) |
| 0.0 | 0.0 | GO:0035381 | purinergic nucleotide receptor activity(GO:0001614) extracellular ATP-gated cation channel activity(GO:0004931) nucleotide receptor activity(GO:0016502) ATP-gated ion channel activity(GO:0035381) purinergic receptor activity(GO:0035586) |
| 0.0 | 0.9 | GO:0004620 | phospholipase activity(GO:0004620) |
| 0.0 | 1.0 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.0 | 0.3 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.6 | PID_RAS_PATHWAY | Regulation of Ras family activation |
| 0.1 | 3.3 | PID_EPHA_FWDPATHWAY | EPHA forward signaling |
| 0.1 | 2.6 | PID_WNT_SIGNALING_PATHWAY | Wnt signaling network |
| 0.1 | 2.4 | PID_EPO_PATHWAY | EPO signaling pathway |
| 0.1 | 2.1 | PID_SYNDECAN_4_PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 1.8 | PID_RET_PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 4.5 | PID_LYSOPHOSPHOLIPID_PATHWAY | LPA receptor mediated events |
| 0.1 | 0.9 | SA_REG_CASCADE_OF_CYCLIN_EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 4.6 | PID_HIF1_TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 3.6 | PID_HDAC_CLASSI_PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 1.6 | ST_MYOCYTE_AD_PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 3.7 | PID_NOTCH_PATHWAY | Notch signaling pathway |
| 0.0 | 1.7 | PID_RETINOIC_ACID_PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 3.9 | PID_MYC_ACTIV_PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 1.5 | SIG_CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 1.0 | PID_INTEGRIN_A4B1_PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 1.4 | PID_FGF_PATHWAY | FGF signaling pathway |
| 0.0 | 0.8 | PID_ARF_3PATHWAY | Arf1 pathway |
| 0.0 | 0.9 | PID_FCER1_PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 1.3 | PID_MYC_REPRESS_PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.6 | PID_REELIN_PATHWAY | Reelin signaling pathway |
| 0.0 | 0.9 | PID_CASPASE_PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.6 | PID_RAC1_PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.2 | PID_BETA_CATENIN_DEG_PATHWAY | Degradation of beta catenin |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.6 | REACTOME_CDC6_ASSOCIATION_WITH_THE_ORC_ORIGIN_COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.2 | 7.4 | REACTOME_PROSTACYCLIN_SIGNALLING_THROUGH_PROSTACYCLIN_RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.2 | 1.4 | REACTOME_FGFR4_LIGAND_BINDING_AND_ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.2 | 2.6 | REACTOME_INHIBITION_OF_VOLTAGE_GATED_CA2_CHANNELS_VIA_GBETA_GAMMA_SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.2 | 2.2 | REACTOME_CHYLOMICRON_MEDIATED_LIPID_TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 6.1 | REACTOME_GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 7.2 | REACTOME_NUCLEAR_RECEPTOR_TRANSCRIPTION_PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 1.7 | REACTOME_CYCLIN_A_B1_ASSOCIATED_EVENTS_DURING_G2_M_TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 1.5 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_LATE_ENDOSOME_MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 3.7 | REACTOME_ACTIVATED_NOTCH1_TRANSMITS_SIGNAL_TO_THE_NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 4.3 | REACTOME_CLASS_B_2_SECRETIN_FAMILY_RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 1.4 | REACTOME_MITOCHONDRIAL_FATTY_ACID_BETA_OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 1.2 | REACTOME_INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.1 | 3.0 | REACTOME_MRNA_3_END_PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.1 | 1.0 | REACTOME_APOPTOSIS_INDUCED_DNA_FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 5.2 | REACTOME_G_ALPHA_I_SIGNALLING_EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 1.3 | REACTOME_SYNTHESIS_OF_GLYCOSYLPHOSPHATIDYLINOSITOL_GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 1.8 | REACTOME_NCAM1_INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 1.5 | REACTOME_O_LINKED_GLYCOSYLATION_OF_MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.5 | REACTOME_P130CAS_LINKAGE_TO_MAPK_SIGNALING_FOR_INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 1.4 | REACTOME_FORMATION_OF_THE_TERNARY_COMPLEX_AND_SUBSEQUENTLY_THE_43S_COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 2.1 | REACTOME_RESPONSE_TO_ELEVATED_PLATELET_CYTOSOLIC_CA2_ | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.9 | REACTOME_NEGATIVE_REGULATORS_OF_RIG_I_MDA5_SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 2.0 | REACTOME_MRNA_SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 1.3 | REACTOME_RESPIRATORY_ELECTRON_TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 1.9 | REACTOME_FACTORS_INVOLVED_IN_MEGAKARYOCYTE_DEVELOPMENT_AND_PLATELET_PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |