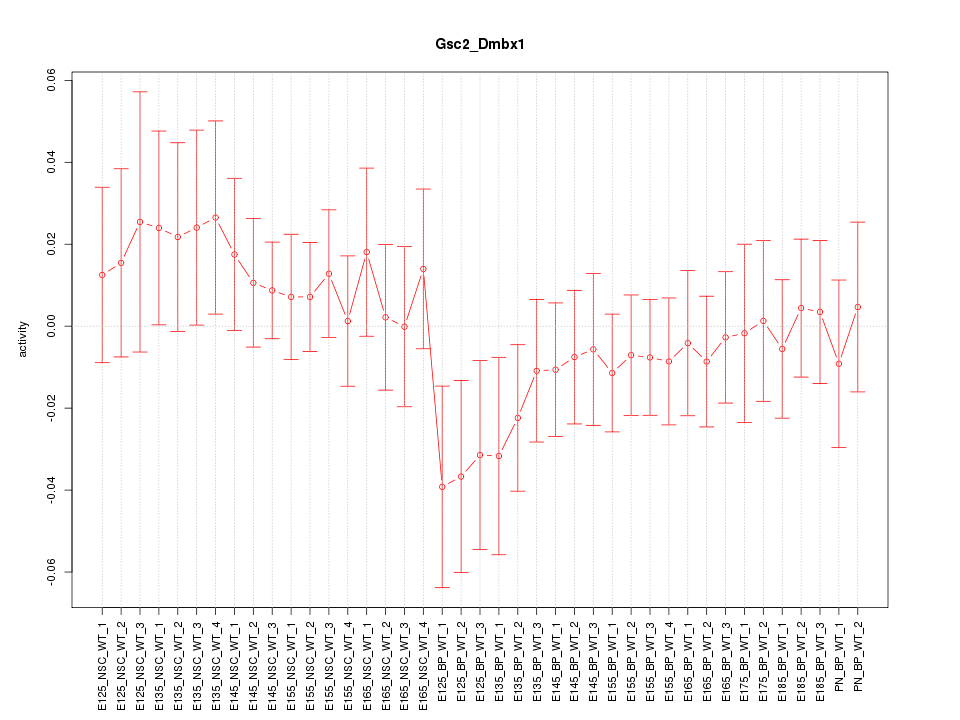
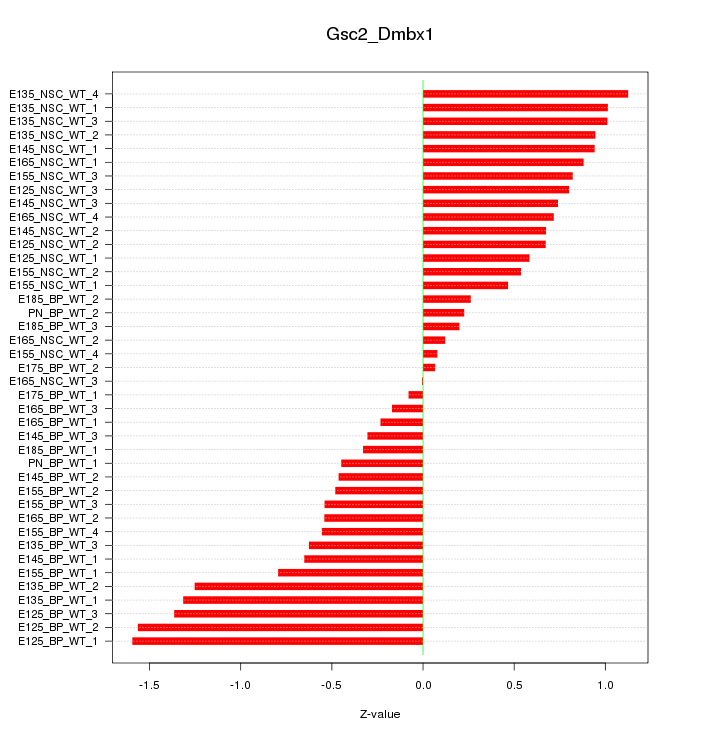
Motif ID: Gsc2_Dmbx1
Z-value: 0.759
Transcription factors associated with Gsc2_Dmbx1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Dmbx1 | ENSMUSG00000028707.9 | Dmbx1 |
| Gsc2 | ENSMUSG00000022738.6 | Gsc2 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Dmbx1 | mm10_v2_chr4_-_115939923_115939928 | 0.53 | 4.2e-04 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 7.4 | GO:2000974 | negative regulation of pro-B cell differentiation(GO:2000974) |
| 1.5 | 8.8 | GO:0006570 | tyrosine metabolic process(GO:0006570) |
| 1.4 | 9.6 | GO:0044838 | cell quiescence(GO:0044838) |
| 1.1 | 3.2 | GO:0032650 | regulation of interleukin-1 alpha production(GO:0032650) positive regulation of interleukin-1 alpha production(GO:0032730) cardiac cell fate determination(GO:0060913) |
| 1.0 | 2.9 | GO:1902524 | interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) positive regulation of protein K48-linked ubiquitination(GO:1902524) |
| 0.8 | 3.0 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.5 | 2.1 | GO:2000705 | regulation of dense core granule biogenesis(GO:2000705) |
| 0.5 | 5.2 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.5 | 4.0 | GO:0002681 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.5 | 2.8 | GO:0086028 | bundle of His cell to Purkinje myocyte signaling(GO:0086028) bundle of His cell action potential(GO:0086043) |
| 0.4 | 1.9 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.3 | 10.8 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.3 | 0.8 | GO:0048687 | positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) |
| 0.3 | 1.8 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.2 | 1.0 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.2 | 5.7 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 1.1 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 0.5 | GO:0036089 | cleavage furrow formation(GO:0036089) |
| 0.1 | 3.2 | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) |
| 0.1 | 1.3 | GO:0090331 | negative regulation of platelet aggregation(GO:0090331) |
| 0.1 | 3.0 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 0.5 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.1 | 0.2 | GO:0001692 | histamine metabolic process(GO:0001692) imidazole-containing compound catabolic process(GO:0052805) |
| 0.0 | 2.7 | GO:0006911 | phagocytosis, engulfment(GO:0006911) |
| 0.0 | 0.2 | GO:0090472 | dibasic protein processing(GO:0090472) |
| 0.0 | 0.5 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 0.6 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.8 | GO:0040034 | regulation of development, heterochronic(GO:0040034) |
| 0.0 | 0.1 | GO:0016056 | photoreceptor cell morphogenesis(GO:0008594) rhodopsin mediated signaling pathway(GO:0016056) post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.0 | 0.9 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 1.7 | GO:0010389 | regulation of G2/M transition of mitotic cell cycle(GO:0010389) |
| 0.0 | 0.4 | GO:0055070 | copper ion homeostasis(GO:0055070) |
| 0.0 | 0.1 | GO:1904778 | regulation of metaphase plate congression(GO:0090235) regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.2 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.7 | GO:1902042 | negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.0 | 0.5 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 1.2 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.4 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 1.9 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 1.0 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.3 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 10.8 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.3 | 8.8 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.3 | 0.8 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.2 | 4.3 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.2 | 11.3 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 3.0 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 4.0 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.1 | 3.0 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 0.5 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 2.7 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 2.8 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 0.5 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 3.2 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.1 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.0 | 0.3 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.2 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 9.6 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 2.7 | 10.8 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.9 | 5.2 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.7 | 8.8 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.4 | 5.7 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.4 | 1.9 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.2 | 1.9 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.2 | 1.3 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.2 | 1.0 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.2 | 2.1 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.1 | 1.0 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.1 | 1.6 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.1 | 1.2 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.1 | 0.4 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.1 | 1.3 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 3.2 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 3.1 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin binding(GO:0030332) |
| 0.1 | 2.8 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 3.0 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.5 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 11.5 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.8 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.5 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.1 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.2 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.5 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 1.2 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.3 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 1.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 5.0 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 0.9 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 0.0 | 0.6 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 9.6 | PID_IL2_STAT5_PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 7.4 | PID_CDC42_PATHWAY | CDC42 signaling events |
| 0.0 | 3.9 | PID_BETA_CATENIN_NUC_PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 1.7 | PID_REG_GR_PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 2.3 | NABA_ECM_REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.4 | REACTOME_SYNTHESIS_SECRETION_AND_INACTIVATION_OF_GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.2 | 9.6 | REACTOME_G1_PHASE | Genes involved in G1 Phase |
| 0.1 | 5.2 | REACTOME_GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 7.4 | REACTOME_NOTCH1_INTRACELLULAR_DOMAIN_REGULATES_TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 1.9 | REACTOME_PEROXISOMAL_LIPID_METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 3.0 | REACTOME_RNA_POL_I_PROMOTER_OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 4.0 | REACTOME_PEPTIDE_CHAIN_ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 1.8 | REACTOME_TRANSCRIPTIONAL_REGULATION_OF_WHITE_ADIPOCYTE_DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 1.6 | REACTOME_ACTIVATION_OF_CHAPERONE_GENES_BY_XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.2 | REACTOME_GAMMA_CARBOXYLATION_TRANSPORT_AND_AMINO_TERMINAL_CLEAVAGE_OF_PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |