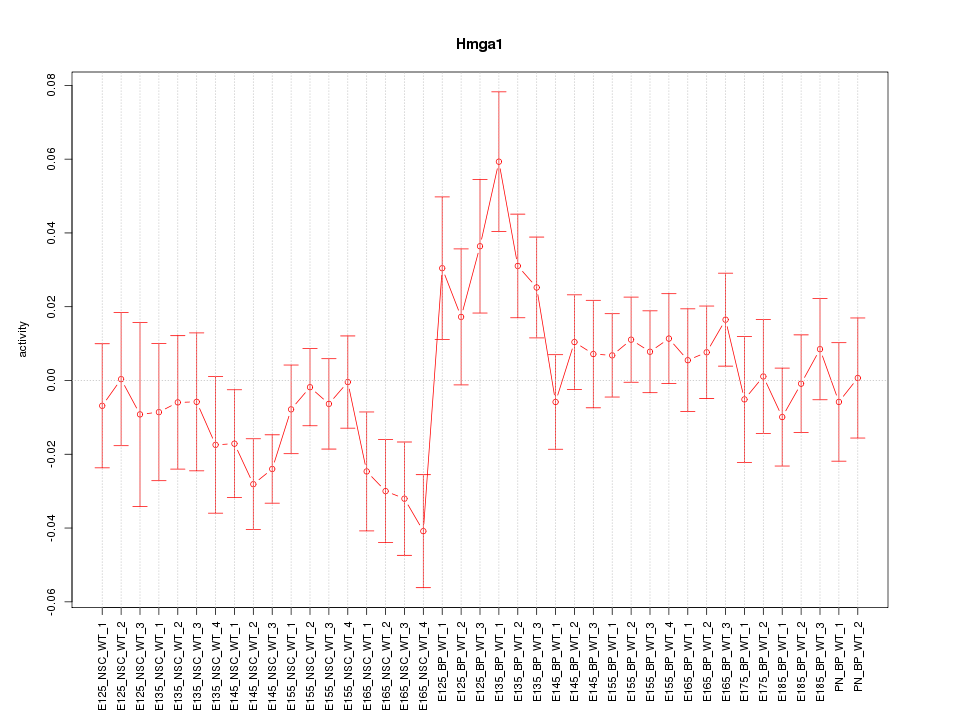
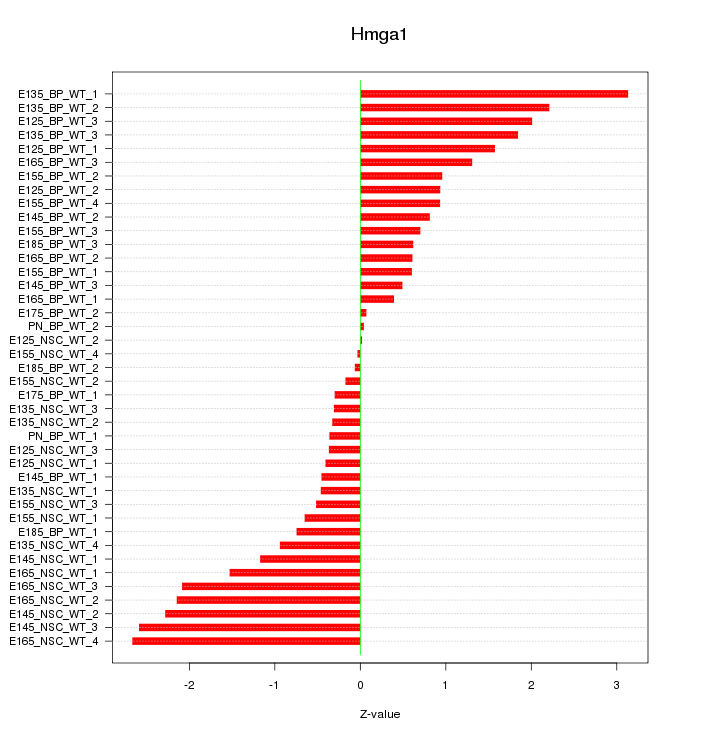
Motif ID: Hmga1
Z-value: 1.275
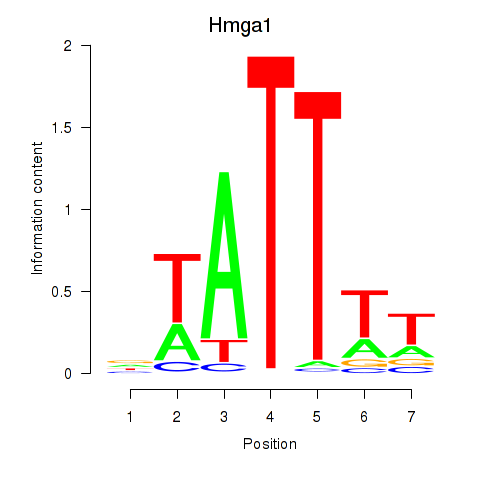
Transcription factors associated with Hmga1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Hmga1 | ENSMUSG00000046711.9 | Hmga1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hmga1 | mm10_v2_chr17_+_27556668_27556690 | 0.06 | 7.0e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.4 | 20.4 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 1.5 | 6.0 | GO:1900623 | positive regulation of keratinocyte proliferation(GO:0010838) regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 1.2 | 3.5 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 1.0 | 4.0 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 1.0 | 3.0 | GO:0003345 | proepicardium cell migration involved in pericardium morphogenesis(GO:0003345) |
| 0.8 | 2.3 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.7 | 2.2 | GO:1902071 | positive regulation of cellular response to hypoxia(GO:1900039) regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 0.7 | 13.3 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.6 | 2.4 | GO:0010511 | regulation of phosphatidylinositol biosynthetic process(GO:0010511) |
| 0.6 | 3.4 | GO:0021773 | striatal medium spiny neuron differentiation(GO:0021773) |
| 0.6 | 2.8 | GO:0060467 | negative regulation of fertilization(GO:0060467) |
| 0.5 | 2.3 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.5 | 2.8 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 0.5 | 2.3 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.4 | 2.7 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) phosphate ion transmembrane transport(GO:0035435) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.4 | 3.8 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.4 | 2.0 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.4 | 2.2 | GO:0045919 | complement activation, alternative pathway(GO:0006957) positive regulation of cytolysis(GO:0045919) |
| 0.4 | 1.5 | GO:0046552 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.4 | 1.5 | GO:0044330 | canonical Wnt signaling pathway involved in positive regulation of wound healing(GO:0044330) lactic acid secretion(GO:0046722) regulation of metanephric cap mesenchymal cell proliferation(GO:0090095) positive regulation of metanephric cap mesenchymal cell proliferation(GO:0090096) |
| 0.4 | 1.4 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 0.3 | 1.0 | GO:0098928 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 0.3 | 1.3 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.3 | 2.0 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.3 | 1.8 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.3 | 1.6 | GO:0032796 | uropod organization(GO:0032796) |
| 0.3 | 1.8 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) regulation of cellular ketone metabolic process by positive regulation of transcription from RNA polymerase II promoter(GO:0072366) |
| 0.3 | 1.0 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.3 | 3.0 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.3 | 1.0 | GO:0019355 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.2 | 1.7 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.2 | 1.3 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.2 | 1.7 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.2 | 0.6 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.2 | 1.0 | GO:0032329 | serine transport(GO:0032329) |
| 0.2 | 6.0 | GO:0007617 | mating behavior(GO:0007617) multi-organism reproductive behavior(GO:0044705) |
| 0.2 | 0.6 | GO:0002182 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.2 | 0.6 | GO:1990927 | negative regulation of membrane invagination(GO:1905154) calcium ion regulated lysosome exocytosis(GO:1990927) |
| 0.2 | 8.5 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.2 | 0.5 | GO:0043323 | positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.2 | 0.7 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.2 | 1.1 | GO:0098734 | protein depalmitoylation(GO:0002084) negative regulation of Golgi to plasma membrane protein transport(GO:0042997) negative regulation of establishment of protein localization to plasma membrane(GO:0090005) macromolecule depalmitoylation(GO:0098734) |
| 0.2 | 0.4 | GO:1903244 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.2 | 0.5 | GO:1902277 | negative regulation of pancreatic amylase secretion(GO:1902277) |
| 0.2 | 0.7 | GO:0046909 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.2 | 1.2 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.2 | 0.9 | GO:0015692 | vanadium ion transport(GO:0015676) lead ion transport(GO:0015692) |
| 0.2 | 0.5 | GO:1900127 | positive regulation of hyaluronan biosynthetic process(GO:1900127) |
| 0.2 | 2.5 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.2 | 1.0 | GO:0002002 | regulation of angiotensin levels in blood(GO:0002002) |
| 0.2 | 0.6 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.2 | 0.9 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.2 | 0.5 | GO:0035799 | ureter maturation(GO:0035799) cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) regulation of DNA demethylation(GO:1901535) negative regulation of DNA demethylation(GO:1901536) |
| 0.2 | 0.3 | GO:0072501 | cellular divalent inorganic anion homeostasis(GO:0072501) |
| 0.1 | 0.9 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.1 | 0.6 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.1 | 0.6 | GO:0060315 | negative regulation of ryanodine-sensitive calcium-release channel activity(GO:0060315) |
| 0.1 | 1.4 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.1 | 0.5 | GO:0036216 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060368) |
| 0.1 | 3.6 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.1 | 0.6 | GO:0072530 | purine-containing compound transmembrane transport(GO:0072530) |
| 0.1 | 0.5 | GO:0090365 | regulation of mRNA modification(GO:0090365) |
| 0.1 | 0.5 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.1 | 0.4 | GO:0021972 | corticospinal neuron axon guidance through spinal cord(GO:0021972) positive regulation of negative chemotaxis(GO:0050924) induction of negative chemotaxis(GO:0050929) negative regulation of mononuclear cell migration(GO:0071676) negative regulation of retinal ganglion cell axon guidance(GO:0090260) |
| 0.1 | 1.6 | GO:0034390 | smooth muscle cell apoptotic process(GO:0034390) regulation of smooth muscle cell apoptotic process(GO:0034391) |
| 0.1 | 1.0 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.1 | 0.3 | GO:0051030 | snRNA transport(GO:0051030) |
| 0.1 | 1.0 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 3.2 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.1 | 0.4 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.1 | 0.3 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.1 | 0.9 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.1 | 0.4 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.1 | 3.5 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.1 | 1.4 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 5.0 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.1 | 1.2 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.1 | 1.9 | GO:0006625 | protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.1 | 1.3 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.1 | 0.6 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.1 | 0.8 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.1 | 3.0 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.1 | 0.4 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 0.5 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.1 | 1.6 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.1 | 0.9 | GO:0006706 | steroid catabolic process(GO:0006706) |
| 0.1 | 0.3 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.1 | 0.7 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.1 | 0.2 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.1 | 0.5 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.1 | 0.4 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.1 | 0.6 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.1 | 4.3 | GO:0060349 | bone morphogenesis(GO:0060349) |
| 0.1 | 5.6 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 0.5 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.1 | 0.9 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.1 | 0.3 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.1 | 0.5 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.1 | 0.3 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.1 | 0.4 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.1 | 0.1 | GO:0071726 | response to diacyl bacterial lipopeptide(GO:0071724) cellular response to diacyl bacterial lipopeptide(GO:0071726) |
| 0.0 | 0.9 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.8 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.0 | 1.1 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 0.1 | GO:0071873 | response to norepinephrine(GO:0071873) |
| 0.0 | 0.2 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 1.3 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.5 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.0 | 0.9 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.8 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 1.6 | GO:0036465 | synaptic vesicle recycling(GO:0036465) |
| 0.0 | 0.6 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.0 | 0.4 | GO:0071549 | cellular response to dexamethasone stimulus(GO:0071549) |
| 0.0 | 0.3 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.2 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.2 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.0 | 1.2 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 0.8 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.0 | 0.4 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 2.1 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.5 | GO:0032402 | melanosome transport(GO:0032402) pigment granule transport(GO:0051904) |
| 0.0 | 1.4 | GO:0030500 | regulation of bone mineralization(GO:0030500) |
| 0.0 | 0.2 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.0 | 1.5 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 0.8 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 | 0.7 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.8 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.5 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.0 | 0.1 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 | 2.3 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 0.5 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.1 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.1 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 | 0.1 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 1.5 | GO:0030308 | negative regulation of cell growth(GO:0030308) |
| 0.0 | 1.8 | GO:0001666 | response to hypoxia(GO:0001666) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.4 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.4 | 3.5 | GO:0000805 | X chromosome(GO:0000805) |
| 0.3 | 1.3 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.3 | 3.4 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.3 | 3.8 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.2 | 4.0 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.2 | 0.7 | GO:0042642 | actomyosin, myosin complex part(GO:0042642) |
| 0.2 | 1.7 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.2 | 0.6 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.2 | 2.4 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.2 | 0.9 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.2 | 1.6 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.2 | 0.5 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.2 | 1.5 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.1 | 1.6 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.1 | 0.5 | GO:0045283 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.1 | 0.6 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.1 | 1.7 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 3.3 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 0.9 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.1 | 0.9 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.1 | 0.5 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.1 | 1.1 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.1 | 0.9 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 1.0 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 0.4 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 0.4 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.1 | 2.6 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.1 | 2.7 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 1.3 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 0.6 | GO:0032009 | early phagosome(GO:0032009) |
| 0.1 | 0.6 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.6 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 2.5 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 3.1 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 3.7 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.0 | 0.9 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 2.2 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 4.1 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 0.3 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.9 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 2.8 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.5 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 1.6 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 11.2 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 1.4 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.6 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.3 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.2 | GO:0098644 | complex of collagen trimers(GO:0098644) |
| 0.0 | 1.1 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 0.4 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.2 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 2.1 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.1 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.5 | GO:0045120 | pronucleus(GO:0045120) |
| 0.0 | 0.1 | GO:0000438 | core TFIIH complex portion of holo TFIIH complex(GO:0000438) |
| 0.0 | 0.3 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.0 | 4.5 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.5 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.9 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 2.2 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 6.5 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.7 | GO:0097223 | sperm part(GO:0097223) |
| 0.0 | 2.3 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.4 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.9 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.9 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.7 | GO:0044452 | nucleolar part(GO:0044452) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 6.0 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 1.7 | 10.2 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 1.0 | 3.0 | GO:0004335 | galactokinase activity(GO:0004335) |
| 0.9 | 2.7 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.8 | 2.4 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.7 | 2.1 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.6 | 2.2 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 0.5 | 2.0 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.4 | 2.5 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.3 | 1.7 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.3 | 4.0 | GO:0043495 | protein anchor(GO:0043495) |
| 0.3 | 2.3 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.3 | 1.0 | GO:0004515 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.2 | 1.6 | GO:0048101 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.2 | 1.7 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.2 | 3.8 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.2 | 2.5 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.2 | 1.0 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.2 | 5.0 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.2 | 2.8 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.2 | 0.9 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.2 | 0.9 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) lead ion transmembrane transporter activity(GO:0015094) vanadium ion transmembrane transporter activity(GO:0015100) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.1 | 1.6 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 0.6 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 4.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 1.7 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 5.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 1.7 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 0.5 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.1 | 0.3 | GO:0001888 | glucuronyl-galactosyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0001888) |
| 0.1 | 22.0 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 1.1 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.6 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.1 | 1.6 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 1.2 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.1 | 0.6 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.1 | 3.2 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 5.2 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 2.5 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 1.0 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 0.6 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.1 | 2.4 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 1.4 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 0.8 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.1 | 1.1 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 0.4 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 0.8 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.1 | 1.0 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 0.5 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 0.8 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 1.2 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 2.3 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 1.0 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.3 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.6 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 3.6 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 2.1 | GO:0098811 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.0 | 0.9 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.9 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.3 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 1.5 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.7 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 1.4 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.7 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.7 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.4 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 2.7 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.3 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 2.1 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.4 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.6 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.6 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.0 | 0.3 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.5 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.7 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.6 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.9 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 3.5 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.0 | 0.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.8 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 0.7 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 1.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.5 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 2.6 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.3 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.5 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.2 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.1 | GO:0004816 | asparagine-tRNA ligase activity(GO:0004816) |
| 0.0 | 0.1 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.0 | 0.5 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.2 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.6 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.8 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.1 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.0 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.0 | 0.6 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.6 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.2 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.5 | GO:0030507 | spectrin binding(GO:0030507) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 10.5 | PID_TCR_CALCIUM_PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 2.9 | PID_FRA_PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 4.0 | PID_CD40_PATHWAY | CD40/CD40L signaling |
| 0.1 | 2.7 | SIG_REGULATION_OF_THE_ACTIN_CYTOSKELETON_BY_RHO_GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 2.0 | NABA_PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 1.8 | PID_HNF3A_PATHWAY | FOXA1 transcription factor network |
| 0.1 | 2.7 | ST_ERK1_ERK2_MAPK_PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 2.3 | PID_HNF3B_PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 2.0 | NABA_COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 6.4 | NABA_ECM_REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.7 | ST_WNT_BETA_CATENIN_PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 1.0 | ST_TUMOR_NECROSIS_FACTOR_PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.5 | PID_ERBB_NETWORK_PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.9 | ST_PHOSPHOINOSITIDE_3_KINASE_PATHWAY | PI3K Pathway |
| 0.0 | 2.8 | PID_CDC42_PATHWAY | CDC42 signaling events |
| 0.0 | 0.6 | PID_NEPHRIN_NEPH1_PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.9 | PID_HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.5 | PID_IL27_PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.8 | PID_AURORA_B_PATHWAY | Aurora B signaling |
| 0.0 | 0.9 | ST_FAS_SIGNALING_PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.5 | PID_CERAMIDE_PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.5 | PID_RAS_PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.4 | PID_GLYPICAN_1PATHWAY | Glypican 1 network |
| 0.0 | 0.8 | PID_RAC1_REG_PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.6 | PID_TRKR_PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.4 | REACTOME_SEROTONIN_RECEPTORS | Genes involved in Serotonin receptors |
| 0.4 | 2.2 | REACTOME_REGULATION_OF_COMPLEMENT_CASCADE | Genes involved in Regulation of Complement cascade |
| 0.4 | 2.5 | REACTOME_ELEVATION_OF_CYTOSOLIC_CA2_LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.4 | 6.0 | REACTOME_HYALURONAN_METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.3 | 4.4 | REACTOME_REGULATION_OF_INSULIN_LIKE_GROWTH_FACTOR_IGF_ACTIVITY_BY_INSULIN_LIKE_GROWTH_FACTOR_BINDING_PROTEINS_IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.2 | 3.8 | REACTOME_GABA_A_RECEPTOR_ACTIVATION | Genes involved in GABA A receptor activation |
| 0.2 | 1.3 | REACTOME_KERATAN_SULFATE_DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.2 | 2.1 | REACTOME_NEF_MEDIATED_DOWNREGULATION_OF_MHC_CLASS_I_COMPLEX_CELL_SURFACE_EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 1.6 | REACTOME_ADVANCED_GLYCOSYLATION_ENDPRODUCT_RECEPTOR_SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 2.5 | REACTOME_ACTIVATION_OF_THE_AP1_FAMILY_OF_TRANSCRIPTION_FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 2.1 | REACTOME_SYNTHESIS_SECRETION_AND_DEACYLATION_OF_GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 2.8 | REACTOME_LYSOSOME_VESICLE_BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.1 | 1.0 | REACTOME_CS_DS_DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 1.7 | REACTOME_N_GLYCAN_ANTENNAE_ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 0.8 | REACTOME_NA_CL_DEPENDENT_NEUROTRANSMITTER_TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 1.7 | REACTOME_HORMONE_SENSITIVE_LIPASE_HSL_MEDIATED_TRIACYLGLYCEROL_HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 2.8 | REACTOME_SMAD2_SMAD3_SMAD4_HETEROTRIMER_REGULATES_TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 1.0 | REACTOME_ACTIVATION_OF_RAC | Genes involved in Activation of Rac |
| 0.1 | 1.6 | REACTOME_CGMP_EFFECTS | Genes involved in cGMP effects |
| 0.1 | 1.4 | REACTOME_RAP1_SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 2.0 | REACTOME_NCAM1_INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 2.4 | REACTOME_AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.5 | REACTOME_ROLE_OF_DCC_IN_REGULATING_APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 1.0 | REACTOME_PHASE1_FUNCTIONALIZATION_OF_COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 0.5 | REACTOME_SHC1_EVENTS_IN_EGFR_SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 2.5 | REACTOME_RESPIRATORY_ELECTRON_TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.7 | REACTOME_INSULIN_SYNTHESIS_AND_PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.9 | REACTOME_ADHERENS_JUNCTIONS_INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 1.3 | REACTOME_ACTIVATION_OF_NF_KAPPAB_IN_B_CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 0.2 | REACTOME_DESTABILIZATION_OF_MRNA_BY_AUF1_HNRNP_D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 1.0 | REACTOME_METABOLISM_OF_VITAMINS_AND_COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.6 | REACTOME_RECYCLING_PATHWAY_OF_L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 2.5 | REACTOME_SIGNALING_BY_RHO_GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.1 | REACTOME_PRE_NOTCH_PROCESSING_IN_GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.1 | REACTOME_IKK_COMPLEX_RECRUITMENT_MEDIATED_BY_RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |