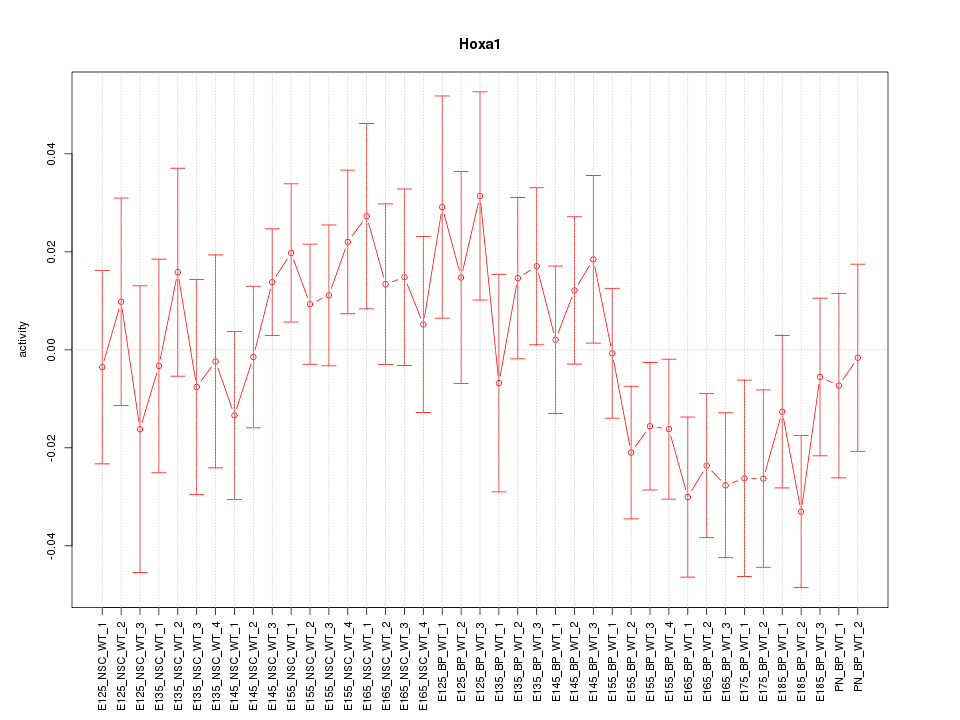
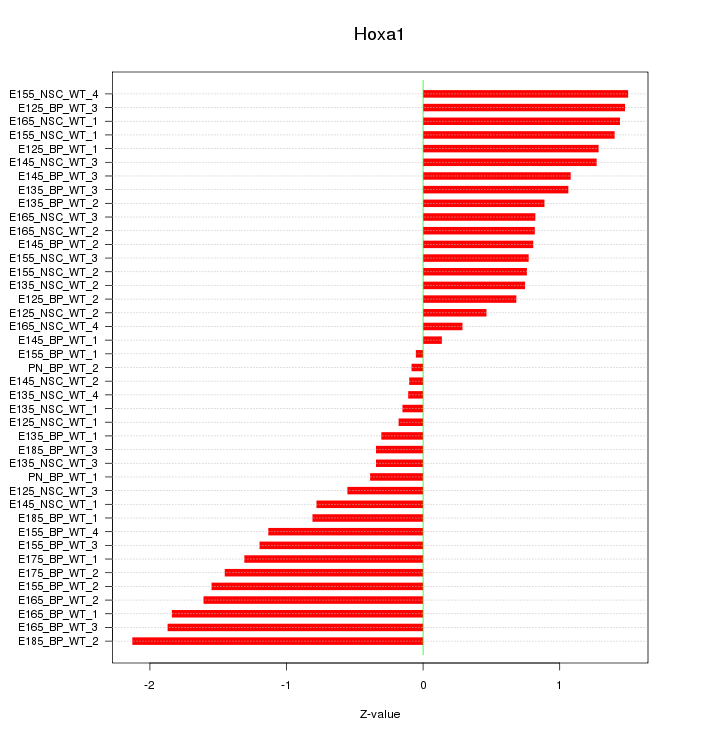
Motif ID: Hoxa1
Z-value: 1.041
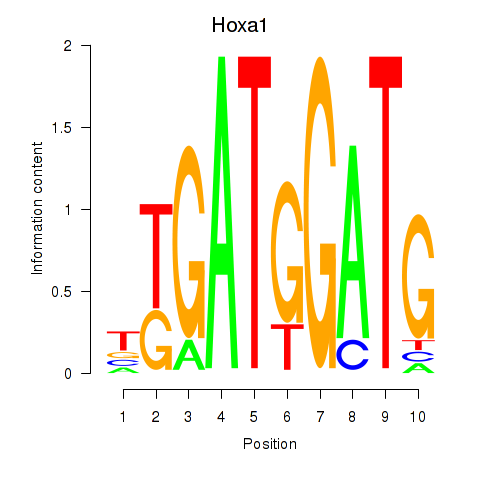
Transcription factors associated with Hoxa1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Hoxa1 | ENSMUSG00000029844.9 | Hoxa1 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 13.9 | GO:1901731 | positive regulation of platelet aggregation(GO:1901731) |
| 1.3 | 4.0 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 0.5 | 3.8 | GO:0033563 | dorsal/ventral axon guidance(GO:0033563) |
| 0.4 | 5.2 | GO:1900121 | negative regulation of receptor binding(GO:1900121) |
| 0.4 | 1.6 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.4 | 4.8 | GO:0033089 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.3 | 0.5 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.2 | 1.7 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.2 | 2.7 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.2 | 2.7 | GO:0042473 | outer ear morphogenesis(GO:0042473) |
| 0.2 | 1.1 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.2 | 1.2 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 0.2 | 2.6 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.2 | 1.3 | GO:2001054 | negative regulation of mesenchymal cell apoptotic process(GO:2001054) |
| 0.1 | 0.3 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.1 | 1.0 | GO:1903847 | regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 0.1 | 0.4 | GO:0072272 | proximal/distal pattern formation involved in metanephric nephron development(GO:0072272) |
| 0.1 | 1.7 | GO:0042711 | maternal behavior(GO:0042711) |
| 0.1 | 0.2 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.1 | 0.5 | GO:0043084 | penile erection(GO:0043084) |
| 0.1 | 0.2 | GO:0051030 | snRNA transport(GO:0051030) |
| 0.0 | 0.9 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 3.6 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.3 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 2.0 | GO:1901381 | positive regulation of potassium ion transmembrane transport(GO:1901381) |
| 0.0 | 1.5 | GO:0009409 | response to cold(GO:0009409) |
| 0.0 | 0.2 | GO:1904800 | regulation of neuron remodeling(GO:1904799) negative regulation of neuron remodeling(GO:1904800) |
| 0.0 | 1.0 | GO:0051968 | positive regulation of synaptic transmission, glutamatergic(GO:0051968) |
| 0.0 | 0.3 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.0 | 0.5 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.0 | 0.4 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.3 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.0 | 0.4 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.6 | GO:0045070 | positive regulation of viral genome replication(GO:0045070) |
| 0.0 | 1.7 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.3 | GO:0001553 | luteinization(GO:0001553) |
| 0.0 | 0.6 | GO:0030947 | regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030947) |
| 0.0 | 0.8 | GO:0007019 | microtubule depolymerization(GO:0007019) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 13.9 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.2 | 1.1 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.2 | 5.3 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 0.4 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.1 | 2.6 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.1 | 1.6 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 3.0 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 3.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.4 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 1.5 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 1.0 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 2.7 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 3.8 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.2 | GO:0035102 | PRC1 complex(GO:0035102) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 4.0 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 1.3 | 13.9 | GO:0015250 | water channel activity(GO:0015250) |
| 0.5 | 3.8 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.4 | 5.2 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.3 | 1.1 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.2 | 1.6 | GO:0031432 | thyroid hormone receptor coactivator activity(GO:0030375) titin binding(GO:0031432) |
| 0.2 | 0.9 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.2 | 1.1 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.1 | 0.3 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.1 | 1.3 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 1.5 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.1 | 2.6 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 1.1 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.3 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 1.4 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 6.2 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 2.7 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 2.0 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.5 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 3.8 | GO:0016779 | nucleotidyltransferase activity(GO:0016779) |
| 0.0 | 0.5 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 3.1 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.4 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.5 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.2 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 1.2 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 2.1 | GO:0016874 | ligase activity(GO:0016874) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.2 | PID_BMP_PATHWAY | BMP receptor signaling |
| 0.1 | 4.8 | PID_NFAT_TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 3.8 | PID_AJDISS_2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 2.7 | PID_FOXM1_PATHWAY | FOXM1 transcription factor network |
| 0.1 | 4.0 | PID_ARF6_TRAFFICKING_PATHWAY | Arf6 trafficking events |
| 0.0 | 1.6 | NABA_BASEMENT_MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 1.0 | PID_EPHRINB_REV_PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.7 | PID_MAPK_TRK_PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 1.5 | PID_AR_PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.4 | PID_SYNDECAN_4_PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 1.3 | NABA_ECM_GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | REACTOME_UNBLOCKING_OF_NMDA_RECEPTOR_GLUTAMATE_BINDING_AND_ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.1 | 9.0 | REACTOME_NETRIN1_SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 1.4 | REACTOME_SIGNAL_REGULATORY_PROTEIN_SIRP_FAMILY_INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 2.7 | REACTOME_CREB_PHOSPHORYLATION_THROUGH_THE_ACTIVATION_OF_CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.1 | 1.1 | REACTOME_FGFR1_LIGAND_BINDING_AND_ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.1 | 1.0 | REACTOME_NEF_MEDIATED_DOWNREGULATION_OF_MHC_CLASS_I_COMPLEX_CELL_SURFACE_EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 1.2 | REACTOME_LYSOSOME_VESICLE_BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 4.0 | REACTOME_PEPTIDE_LIGAND_BINDING_RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.5 | REACTOME_RECEPTOR_LIGAND_BINDING_INITIATES_THE_SECOND_PROTEOLYTIC_CLEAVAGE_OF_NOTCH_RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 2.6 | REACTOME_G_ALPHA_I_SIGNALLING_EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.4 | REACTOME_NEPHRIN_INTERACTIONS | Genes involved in Nephrin interactions |