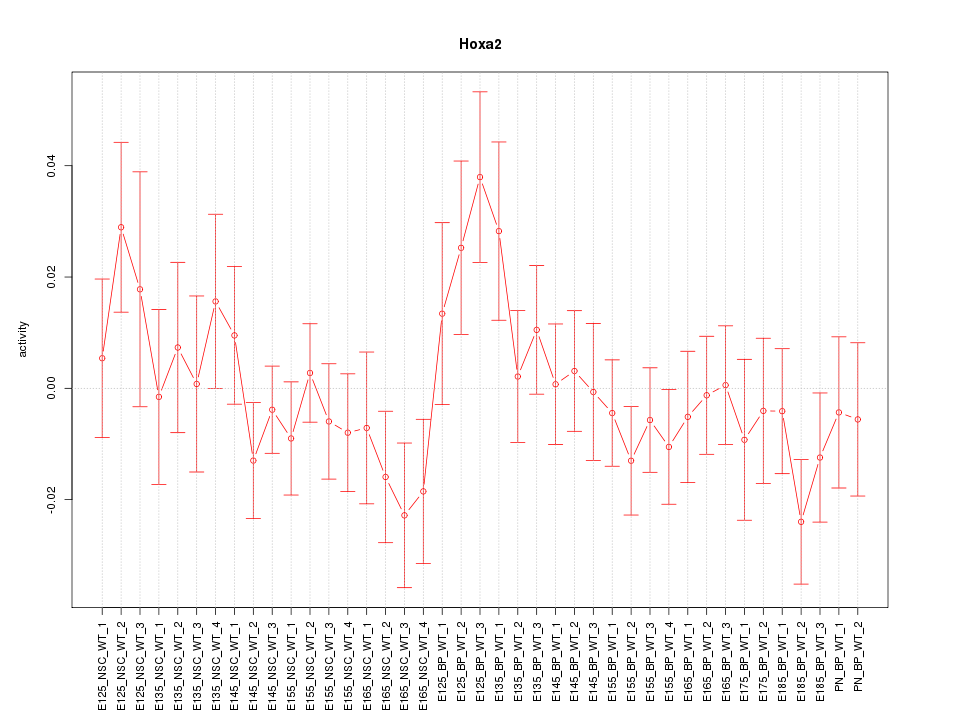
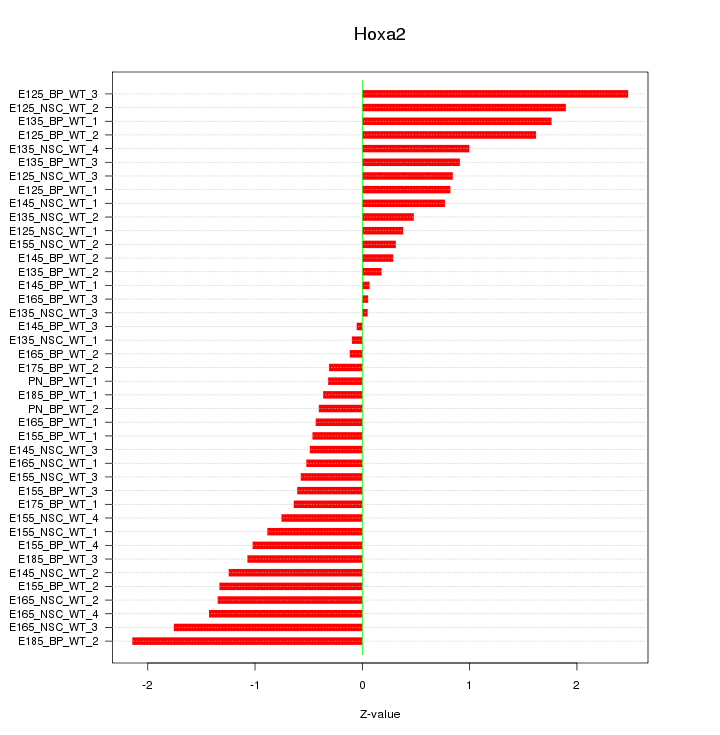
Motif ID: Hoxa2
Z-value: 0.998

Transcription factors associated with Hoxa2:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Hoxa2 | ENSMUSG00000014704.8 | Hoxa2 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 21.7 | GO:1902715 | positive regulation of interferon-gamma secretion(GO:1902715) |
| 1.8 | 12.6 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 1.3 | 3.9 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 1.0 | 3.1 | GO:0001869 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 1.0 | 2.9 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.9 | 4.3 | GO:0046864 | retinol transport(GO:0034633) isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) alveolar primary septum development(GO:0061143) |
| 0.7 | 2.2 | GO:1902713 | regulation of interferon-gamma secretion(GO:1902713) |
| 0.7 | 2.1 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.6 | 0.6 | GO:2000277 | positive regulation of oxidative phosphorylation uncoupler activity(GO:2000277) |
| 0.6 | 2.4 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.6 | 2.9 | GO:0032901 | positive regulation of neurotrophin production(GO:0032901) |
| 0.6 | 2.3 | GO:0061743 | motor learning(GO:0061743) |
| 0.6 | 2.3 | GO:0046368 | GDP-L-fucose metabolic process(GO:0046368) |
| 0.5 | 1.6 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 0.5 | 2.5 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.5 | 1.4 | GO:0002636 | positive regulation of germinal center formation(GO:0002636) |
| 0.4 | 1.3 | GO:0019343 | cysteine biosynthetic process via cystathionine(GO:0019343) cysteine biosynthetic process(GO:0019344) |
| 0.4 | 1.6 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.4 | 2.8 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.4 | 2.4 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.4 | 1.2 | GO:0051030 | snRNA transport(GO:0051030) |
| 0.4 | 1.9 | GO:0072235 | distal convoluted tubule development(GO:0072025) metanephric distal convoluted tubule development(GO:0072221) metanephric distal tubule development(GO:0072235) |
| 0.4 | 0.7 | GO:0060737 | prostate epithelial cord elongation(GO:0060523) prostate gland morphogenetic growth(GO:0060737) |
| 0.4 | 1.4 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) negative regulation of T-helper 17 type immune response(GO:2000317) regulation of T-helper 17 cell differentiation(GO:2000319) negative regulation of T-helper 17 cell differentiation(GO:2000320) |
| 0.4 | 3.2 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.3 | 1.4 | GO:0051643 | endoplasmic reticulum localization(GO:0051643) |
| 0.3 | 1.0 | GO:1904959 | regulation of electron carrier activity(GO:1904732) regulation of cytochrome-c oxidase activity(GO:1904959) |
| 0.3 | 1.0 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.3 | 0.9 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.3 | 2.4 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.3 | 2.6 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.3 | 1.7 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.3 | 1.1 | GO:0000957 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.3 | 1.9 | GO:0033563 | dorsal/ventral axon guidance(GO:0033563) |
| 0.3 | 1.3 | GO:0072103 | renal system vasculature morphogenesis(GO:0061438) kidney vasculature morphogenesis(GO:0061439) glomerulus vasculature morphogenesis(GO:0072103) glomerular capillary formation(GO:0072104) |
| 0.3 | 2.6 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.3 | 2.0 | GO:0070474 | positive regulation of uterine smooth muscle contraction(GO:0070474) |
| 0.3 | 1.0 | GO:1902340 | telomeric heterochromatin assembly(GO:0031509) negative regulation of chromosome condensation(GO:1902340) |
| 0.2 | 0.7 | GO:0060672 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.2 | 1.9 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.2 | 0.7 | GO:0016132 | brassinosteroid metabolic process(GO:0016131) brassinosteroid biosynthetic process(GO:0016132) |
| 0.2 | 0.7 | GO:2000224 | regulation of testosterone biosynthetic process(GO:2000224) |
| 0.2 | 2.3 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.2 | 6.7 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.2 | 0.7 | GO:0035790 | platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) |
| 0.2 | 0.6 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.2 | 1.1 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.2 | 0.5 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.2 | 0.5 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.2 | 1.1 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.2 | 1.4 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.2 | 0.5 | GO:0018065 | protein-cofactor linkage(GO:0018065) |
| 0.2 | 0.5 | GO:1904578 | response to thapsigargin(GO:1904578) cellular response to thapsigargin(GO:1904579) |
| 0.1 | 1.2 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.1 | 1.0 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.1 | 0.4 | GO:0009446 | putrescine biosynthetic process(GO:0009446) |
| 0.1 | 1.8 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.1 | 0.4 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.1 | 1.2 | GO:0032693 | negative regulation of interleukin-10 production(GO:0032693) |
| 0.1 | 1.0 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.1 | 1.8 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.1 | 1.6 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 0.1 | 1.3 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.1 | 0.5 | GO:0006244 | pyrimidine nucleotide catabolic process(GO:0006244) pyrimidine deoxyribonucleotide catabolic process(GO:0009223) |
| 0.1 | 1.9 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.1 | 0.4 | GO:0019401 | alditol biosynthetic process(GO:0019401) |
| 0.1 | 1.2 | GO:0060484 | lung-associated mesenchyme development(GO:0060484) |
| 0.1 | 0.5 | GO:0099527 | postsynapse to nucleus signaling pathway(GO:0099527) |
| 0.1 | 1.4 | GO:1900121 | negative regulation of receptor binding(GO:1900121) |
| 0.1 | 2.4 | GO:0035036 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) |
| 0.1 | 0.4 | GO:0010796 | regulation of multivesicular body size(GO:0010796) |
| 0.1 | 2.2 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 0.5 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 | 2.3 | GO:0043486 | histone exchange(GO:0043486) |
| 0.1 | 0.4 | GO:1903031 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.1 | 1.1 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 2.4 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.1 | 0.8 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 3.4 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.1 | 0.5 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 0.6 | GO:0060837 | blood vessel endothelial cell differentiation(GO:0060837) |
| 0.1 | 0.4 | GO:0035937 | estrogen secretion(GO:0035937) estradiol secretion(GO:0035938) regulation of estrogen secretion(GO:2000861) regulation of estradiol secretion(GO:2000864) |
| 0.1 | 1.9 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 0.6 | GO:0090234 | regulation of centromere complex assembly(GO:0090230) regulation of kinetochore assembly(GO:0090234) |
| 0.1 | 0.9 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.1 | 0.3 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.1 | 0.9 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.1 | 0.4 | GO:2001184 | regulation of interleukin-12 secretion(GO:2001182) positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.1 | 1.0 | GO:0009125 | nucleoside monophosphate catabolic process(GO:0009125) |
| 0.1 | 4.6 | GO:0008543 | fibroblast growth factor receptor signaling pathway(GO:0008543) |
| 0.1 | 0.3 | GO:0046898 | response to cycloheximide(GO:0046898) |
| 0.1 | 0.3 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.1 | 6.0 | GO:0016525 | negative regulation of angiogenesis(GO:0016525) |
| 0.1 | 0.4 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.1 | 0.8 | GO:0001711 | endodermal cell fate commitment(GO:0001711) |
| 0.1 | 1.1 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.1 | 0.3 | GO:0060414 | response to potassium ion(GO:0035864) cellular response to potassium ion(GO:0035865) aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.1 | 0.9 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 1.2 | GO:0045830 | positive regulation of isotype switching(GO:0045830) |
| 0.1 | 0.9 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.1 | 0.8 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.1 | 1.2 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.1 | 0.7 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 1.1 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 0.5 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.1 | 0.3 | GO:0018002 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.1 | 0.8 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.1 | 0.8 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 0.0 | 0.2 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.0 | 0.2 | GO:0010636 | positive regulation of mitochondrial fusion(GO:0010636) |
| 0.0 | 1.0 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.0 | 0.2 | GO:0021886 | hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) |
| 0.0 | 1.4 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.4 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.3 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.0 | 1.2 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.7 | GO:0019430 | removal of superoxide radicals(GO:0019430) |
| 0.0 | 0.2 | GO:0070813 | hydrogen sulfide metabolic process(GO:0070813) |
| 0.0 | 0.5 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 0.2 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.2 | GO:0060715 | syncytiotrophoblast cell differentiation involved in labyrinthine layer development(GO:0060715) |
| 0.0 | 0.7 | GO:2000269 | regulation of fibroblast apoptotic process(GO:2000269) |
| 0.0 | 0.8 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.0 | 2.0 | GO:0007338 | single fertilization(GO:0007338) |
| 0.0 | 0.7 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 3.8 | GO:0007052 | mitotic spindle organization(GO:0007052) |
| 0.0 | 2.4 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.3 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.3 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 1.0 | GO:0009566 | fertilization(GO:0009566) |
| 0.0 | 1.3 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.2 | GO:0014052 | regulation of gamma-aminobutyric acid secretion(GO:0014052) |
| 0.0 | 0.2 | GO:0039702 | cell separation after cytokinesis(GO:0000920) viral budding via host ESCRT complex(GO:0039702) |
| 0.0 | 0.9 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.0 | 0.1 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.3 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 | 0.3 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.0 | 0.1 | GO:0000076 | DNA replication checkpoint(GO:0000076) |
| 0.0 | 0.9 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.5 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.3 | GO:0051984 | positive regulation of chromosome segregation(GO:0051984) |
| 0.0 | 0.5 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 0.3 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 2.2 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.3 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.0 | 0.7 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.2 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.9 | GO:0043392 | negative regulation of DNA binding(GO:0043392) |
| 0.0 | 0.7 | GO:0002053 | positive regulation of mesenchymal cell proliferation(GO:0002053) |
| 0.0 | 0.8 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.5 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.0 | 0.4 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.4 | GO:0045070 | positive regulation of viral genome replication(GO:0045070) |
| 0.0 | 1.2 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.5 | GO:0006896 | Golgi to vacuole transport(GO:0006896) |
| 0.0 | 0.2 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.3 | GO:0042219 | cellular modified amino acid catabolic process(GO:0042219) |
| 0.0 | 0.8 | GO:0032435 | negative regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032435) |
| 0.0 | 0.2 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.0 | 0.3 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 0.1 | GO:0010826 | negative regulation of centrosome duplication(GO:0010826) negative regulation of centrosome cycle(GO:0046606) |
| 0.0 | 0.1 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 0.1 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.0 | 0.1 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.7 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.4 | GO:0002011 | morphogenesis of an epithelial sheet(GO:0002011) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 6.1 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.8 | 2.4 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.5 | 3.2 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.5 | 2.6 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.4 | 3.9 | GO:0000805 | X chromosome(GO:0000805) |
| 0.3 | 1.9 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.3 | 2.7 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.2 | 1.2 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.2 | 1.4 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.2 | 4.6 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.2 | 0.8 | GO:0002111 | BRCA2-BRAF35 complex(GO:0002111) |
| 0.2 | 1.4 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.1 | 0.3 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.1 | 0.6 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.1 | 0.4 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.1 | 1.3 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 0.9 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.1 | 4.5 | GO:0097546 | ciliary base(GO:0097546) |
| 0.1 | 0.7 | GO:0000938 | GARP complex(GO:0000938) |
| 0.1 | 1.4 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 0.7 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.1 | 0.9 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.1 | 1.4 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 0.9 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 0.5 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.1 | 0.7 | GO:0089701 | U2AF(GO:0089701) |
| 0.1 | 2.5 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 0.8 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.1 | 1.1 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.1 | 0.4 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.1 | 2.1 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 2.1 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 0.3 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.1 | 0.9 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 0.7 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 1.7 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 0.8 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.1 | 0.7 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.1 | 1.0 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 2.0 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 0.2 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.0 | 4.2 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 1.0 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.3 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 1.2 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 3.1 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 1.5 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.7 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 3.2 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 1.4 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.3 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.7 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 0.3 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.3 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.5 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 2.7 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.4 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.4 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.1 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 0.3 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.0 | 1.3 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.4 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 1.1 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.4 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 1.4 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.9 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.8 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.6 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.8 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.3 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 3.0 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 1.0 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.7 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.4 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 1.2 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 2.2 | GO:0044297 | cell body(GO:0044297) |
| 0.0 | 4.8 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.0 | 1.4 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 0.6 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.4 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 1.4 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.0 | 1.0 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 0.4 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.1 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.3 | GO:0032040 | small-subunit processome(GO:0032040) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 24.9 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 1.3 | 3.9 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.6 | 1.9 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.5 | 2.4 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.4 | 1.3 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.4 | 2.5 | GO:0051429 | corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.4 | 2.0 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.4 | 1.8 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.4 | 2.9 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.4 | 2.2 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.3 | 1.4 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.3 | 1.4 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.3 | 0.9 | GO:0031750 | D3 dopamine receptor binding(GO:0031750) |
| 0.3 | 1.4 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.2 | 0.7 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.2 | 1.9 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.2 | 1.0 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.2 | 0.7 | GO:0009918 | sterol delta7 reductase activity(GO:0009918) 7-dehydrocholesterol reductase activity(GO:0047598) |
| 0.2 | 3.3 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.2 | 1.2 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.2 | 0.8 | GO:1990269 | RNA polymerase II C-terminal domain phosphoserine binding(GO:1990269) |
| 0.2 | 5.8 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.2 | 0.5 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.2 | 0.5 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.2 | 0.7 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.2 | 1.3 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.2 | 3.1 | GO:0001848 | complement binding(GO:0001848) |
| 0.2 | 1.2 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.1 | 1.0 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.1 | 1.3 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 0.4 | GO:0046592 | polyamine oxidase activity(GO:0046592) spermine:oxygen oxidoreductase (spermidine-forming) activity(GO:0052901) |
| 0.1 | 0.4 | GO:0047751 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) enone reductase activity(GO:0035671) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.1 | 4.3 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.1 | 0.5 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.1 | 3.0 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 0.9 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.1 | 0.9 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.1 | 1.8 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 1.4 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.1 | 0.5 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 4.6 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 1.1 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 2.0 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.1 | 0.4 | GO:0008967 | phosphoglycolate phosphatase activity(GO:0008967) |
| 0.1 | 0.3 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.1 | 1.2 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 0.4 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.1 | 0.3 | GO:0016880 | acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.1 | 1.4 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.1 | 2.5 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 0.8 | GO:0016004 | phospholipase activator activity(GO:0016004) lipase activator activity(GO:0060229) |
| 0.1 | 2.4 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.1 | 0.5 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.1 | 2.2 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 0.9 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.2 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.1 | 0.8 | GO:0015924 | mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.1 | 0.3 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.1 | 3.4 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 0.3 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.1 | 0.5 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.1 | 0.5 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.1 | 0.5 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.1 | 0.6 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.1 | 1.4 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.1 | 0.5 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.1 | 0.3 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 1.0 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 2.4 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.1 | 0.3 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.1 | 0.3 | GO:1990190 | peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.1 | 0.7 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.1 | 1.1 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.2 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) lead ion transmembrane transporter activity(GO:0015094) vanadium ion transmembrane transporter activity(GO:0015100) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.0 | 1.0 | GO:0016799 | hydrolase activity, hydrolyzing N-glycosyl compounds(GO:0016799) |
| 0.0 | 0.3 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) |
| 0.0 | 1.0 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.6 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.2 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.0 | 3.0 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.8 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 1.6 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 0.5 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.3 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.9 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.3 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.4 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 0.2 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.6 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 4.0 | GO:0016779 | nucleotidyltransferase activity(GO:0016779) |
| 0.0 | 0.8 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.5 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.1 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.8 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 2.0 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.4 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.8 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.2 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.5 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.2 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.4 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.7 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 2.7 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.0 | 0.2 | GO:0016701 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen(GO:0016701) oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.2 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.4 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 0.1 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.4 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 1.6 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 1.8 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 1.1 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.0 | 1.0 | GO:0043178 | alcohol binding(GO:0043178) |
| 0.0 | 0.1 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.0 | GO:0051747 | cytosine C-5 DNA demethylase activity(GO:0051747) |
| 0.0 | 0.8 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 1.8 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 20.9 | PID_RAS_PATHWAY | Regulation of Ras family activation |
| 0.2 | 6.1 | NABA_PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 2.4 | PID_RANBP2_PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 0.5 | PID_PI3KCI_AKT_PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.1 | 1.8 | PID_SMAD2_3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 2.1 | PID_WNT_SIGNALING_PATHWAY | Wnt signaling network |
| 0.1 | 1.4 | PID_IL27_PATHWAY | IL27-mediated signaling events |
| 0.0 | 3.1 | PID_PLK1_PATHWAY | PLK1 signaling events |
| 0.0 | 1.8 | PID_P38_MK2_PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.5 | PID_FCER1_PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 1.1 | PID_MYC_PATHWAY | C-MYC pathway |
| 0.0 | 0.5 | PID_MAPK_TRK_PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 1.3 | PID_P38_ALPHA_BETA_PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 1.4 | PID_BMP_PATHWAY | BMP receptor signaling |
| 0.0 | 0.5 | PID_FOXM1_PATHWAY | FOXM1 transcription factor network |
| 0.0 | 2.3 | PID_ERA_GENOMIC_PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 1.2 | PID_HNF3B_PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.6 | PID_HIF1A_PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.5 | PID_BETA_CATENIN_DEG_PATHWAY | Degradation of beta catenin |
| 0.0 | 1.3 | PID_AJDISS_2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 1.7 | PID_NOTCH_PATHWAY | Notch signaling pathway |
| 0.0 | 0.4 | PID_INTEGRIN_A9B1_PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.7 | PID_BARD1_PATHWAY | BARD1 signaling events |
| 0.0 | 0.3 | SA_PROGRAMMED_CELL_DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.5 | SIG_CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.4 | PID_NCADHERIN_PATHWAY | N-cadherin signaling events |
| 0.0 | 0.4 | PID_RAC1_PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.3 | SIG_REGULATION_OF_THE_ACTIN_CYTOSKELETON_BY_RHO_GTPASES | Genes related to regulation of the actin cytoskeleton |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 22.4 | REACTOME_RAP1_SIGNALLING | Genes involved in Rap1 signalling |
| 0.3 | 3.1 | REACTOME_INTRINSIC_PATHWAY | Genes involved in Intrinsic Pathway |
| 0.2 | 3.0 | REACTOME_FGFR1_LIGAND_BINDING_AND_ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.2 | 4.4 | REACTOME_GLUTAMATE_NEUROTRANSMITTER_RELEASE_CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.2 | 1.8 | REACTOME_REMOVAL_OF_THE_FLAP_INTERMEDIATE_FROM_THE_C_STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.1 | 2.6 | REACTOME_PRE_NOTCH_PROCESSING_IN_GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 1.6 | REACTOME_POST_CHAPERONIN_TUBULIN_FOLDING_PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 0.4 | REACTOME_ANDROGEN_BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.1 | 8.6 | REACTOME_MITOTIC_PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 2.0 | REACTOME_METAL_ION_SLC_TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 3.6 | REACTOME_GLUCAGON_SIGNALING_IN_METABOLIC_REGULATION | Genes involved in Glucagon signaling in metabolic regulation |
| 0.1 | 2.3 | REACTOME_INTERACTION_BETWEEN_L1_AND_ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 1.6 | REACTOME_CELL_EXTRACELLULAR_MATRIX_INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 1.5 | REACTOME_SULFUR_AMINO_ACID_METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 2.0 | REACTOME_INTERACTIONS_OF_VPR_WITH_HOST_CELLULAR_PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.1 | 1.3 | REACTOME_KINESINS | Genes involved in Kinesins |
| 0.1 | 3.0 | REACTOME_ACTIVATION_OF_CHAPERONE_GENES_BY_XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 2.7 | REACTOME_RNA_POL_I_PROMOTER_OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 1.4 | REACTOME_IL_RECEPTOR_SHC_SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 1.2 | REACTOME_METABOLISM_OF_NON_CODING_RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 4.0 | REACTOME_RESPIRATORY_ELECTRON_TRANSPORT_ATP_SYNTHESIS_BY_CHEMIOSMOTIC_COUPLING_AND_HEAT_PRODUCTION_BY_UNCOUPLING_PROTEINS_ | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.0 | 0.4 | REACTOME_METABOLISM_OF_POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 1.0 | REACTOME_CHOLESTEROL_BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 1.9 | REACTOME_ANTIVIRAL_MECHANISM_BY_IFN_STIMULATED_GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.5 | REACTOME_SIGNALLING_TO_P38_VIA_RIT_AND_RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 0.9 | REACTOME_STRIATED_MUSCLE_CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.4 | REACTOME_REGULATION_OF_WATER_BALANCE_BY_RENAL_AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 0.7 | REACTOME_FANCONI_ANEMIA_PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 2.9 | REACTOME_G_ALPHA_I_SIGNALLING_EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.9 | REACTOME_ABC_FAMILY_PROTEINS_MEDIATED_TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.4 | REACTOME_P38MAPK_EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.6 | REACTOME_OTHER_SEMAPHORIN_INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.3 | REACTOME_SYNTHESIS_SECRETION_AND_INACTIVATION_OF_GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.3 | REACTOME_INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 1.0 | REACTOME_CLASS_B_2_SECRETIN_FAMILY_RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.5 | REACTOME_BASIGIN_INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.7 | REACTOME_NUCLEAR_SIGNALING_BY_ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.9 | REACTOME_MITOCHONDRIAL_PROTEIN_IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.9 | REACTOME_AUTODEGRADATION_OF_THE_E3_UBIQUITIN_LIGASE_COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.0 | 1.1 | REACTOME_FACTORS_INVOLVED_IN_MEGAKARYOCYTE_DEVELOPMENT_AND_PLATELET_PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.3 | REACTOME_AMINO_ACID_SYNTHESIS_AND_INTERCONVERSION_TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.3 | REACTOME_DOWNREGULATION_OF_SMAD2_3_SMAD4_TRANSCRIPTIONAL_ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.8 | REACTOME_TRANSCRIPTIONAL_REGULATION_OF_WHITE_ADIPOCYTE_DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.2 | REACTOME_N_GLYCAN_ANTENNAE_ELONGATION_IN_THE_MEDIAL_TRANS_GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |