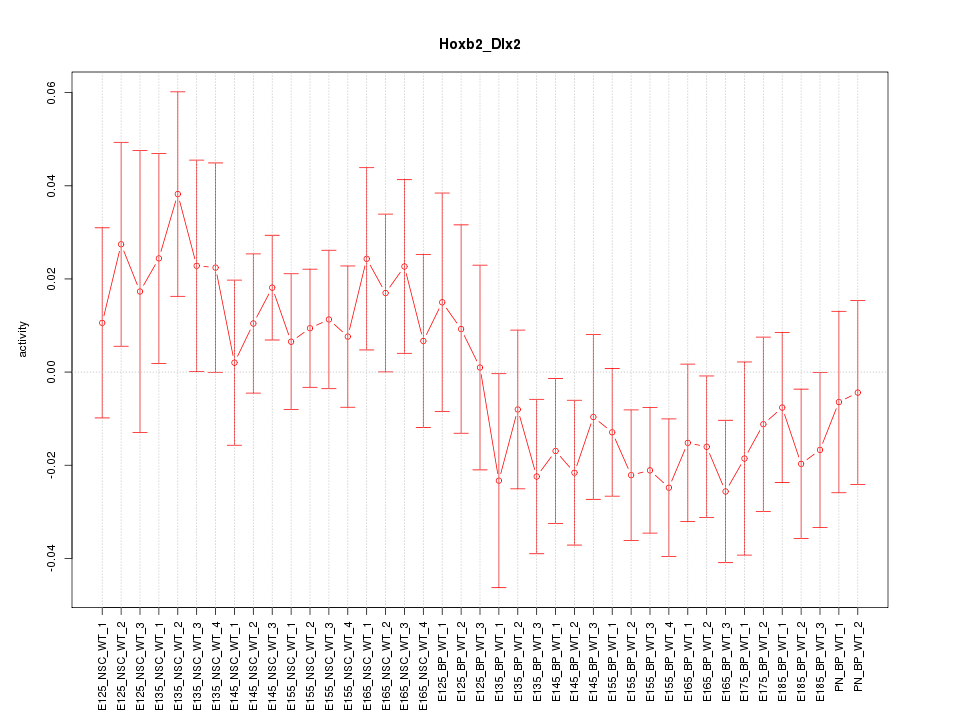
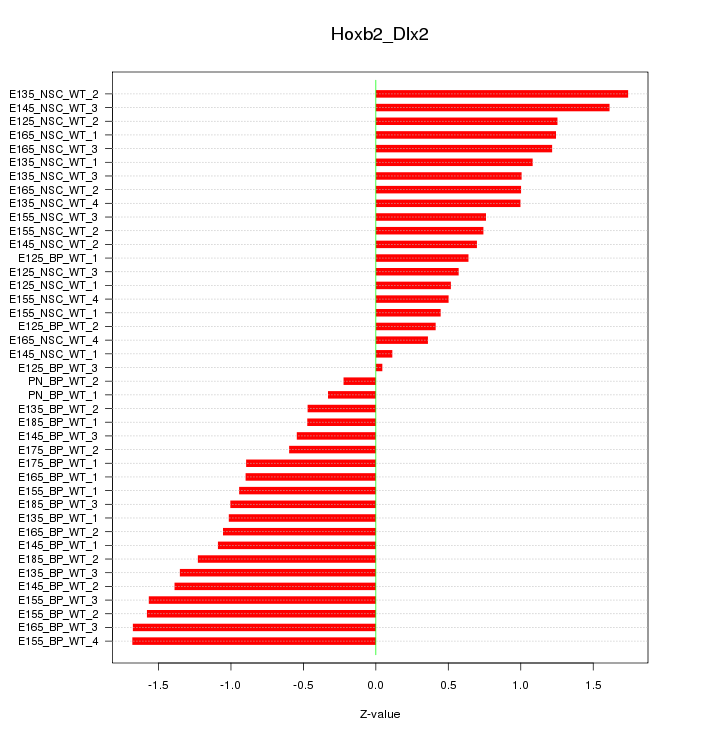
Motif ID: Hoxb2_Dlx2
Z-value: 1.009
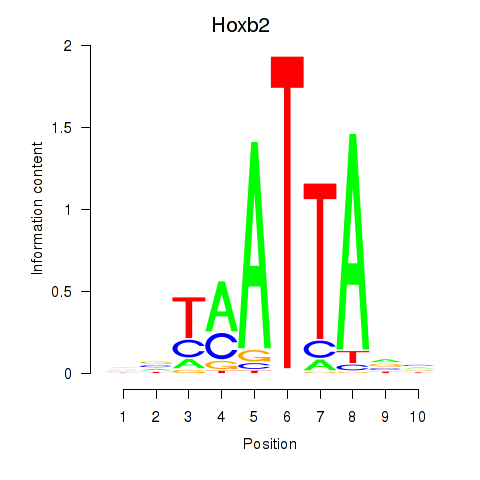
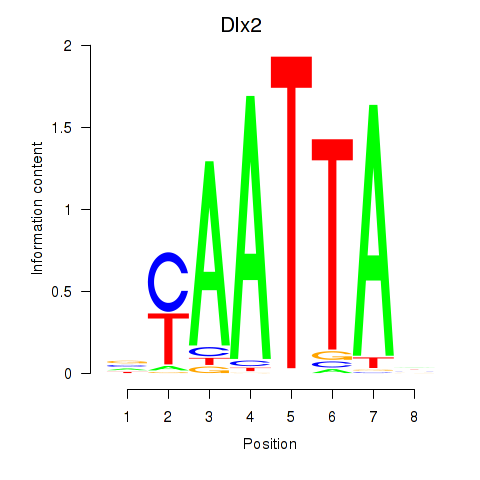
Transcription factors associated with Hoxb2_Dlx2:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Dlx2 | ENSMUSG00000023391.7 | Dlx2 |
| Hoxb2 | ENSMUSG00000075588.5 | Hoxb2 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Dlx2 | mm10_v2_chr2_-_71546745_71546758 | 0.88 | 3.2e-14 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.4 | 25.3 | GO:0061075 | cerebral cortex GABAergic interneuron fate commitment(GO:0021893) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) positive regulation of amacrine cell differentiation(GO:1902871) |
| 3.3 | 13.1 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 2.1 | 8.3 | GO:0046552 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 1.8 | 5.5 | GO:0061357 | Wnt protein secretion(GO:0061355) regulation of Wnt protein secretion(GO:0061356) positive regulation of Wnt protein secretion(GO:0061357) |
| 1.5 | 10.3 | GO:0019532 | oxalate transport(GO:0019532) |
| 1.4 | 15.6 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 1.3 | 5.1 | GO:0061743 | motor learning(GO:0061743) |
| 1.2 | 7.2 | GO:0006570 | tyrosine metabolic process(GO:0006570) |
| 1.2 | 3.6 | GO:0060166 | olfactory pit development(GO:0060166) |
| 1.1 | 7.9 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 1.1 | 4.4 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.9 | 1.9 | GO:1904956 | regulation of midbrain dopaminergic neuron differentiation(GO:1904956) |
| 0.9 | 7.5 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 0.9 | 7.3 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.9 | 4.6 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.8 | 2.3 | GO:0038044 | negative regulation of receptor biosynthetic process(GO:0010871) transforming growth factor-beta secretion(GO:0038044) |
| 0.6 | 1.8 | GO:0060023 | soft palate development(GO:0060023) |
| 0.6 | 1.8 | GO:0090274 | positive regulation of somatostatin secretion(GO:0090274) |
| 0.6 | 1.7 | GO:0019343 | cysteine biosynthetic process via cystathionine(GO:0019343) cysteine biosynthetic process(GO:0019344) |
| 0.5 | 1.6 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) positive regulation of catagen(GO:0051795) frontal suture morphogenesis(GO:0060364) |
| 0.5 | 2.0 | GO:0021648 | zygotic determination of anterior/posterior axis, embryo(GO:0007354) cranial nerve formation(GO:0021603) vestibulocochlear nerve morphogenesis(GO:0021648) |
| 0.5 | 0.5 | GO:2000040 | regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.5 | 1.9 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.5 | 1.0 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.5 | 2.9 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.5 | 2.9 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.5 | 7.2 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.4 | 9.2 | GO:0007530 | sex determination(GO:0007530) |
| 0.4 | 1.3 | GO:0090425 | hepatocyte cell migration(GO:0002194) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 0.4 | 0.4 | GO:0072106 | regulation of ureteric bud formation(GO:0072106) positive regulation of ureteric bud formation(GO:0072107) |
| 0.4 | 2.5 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.4 | 1.9 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.3 | 1.0 | GO:0072204 | cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 0.3 | 1.0 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.3 | 1.9 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.3 | 1.2 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.3 | 0.9 | GO:0045113 | regulation of integrin biosynthetic process(GO:0045113) |
| 0.3 | 2.5 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.3 | 2.7 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.3 | 1.3 | GO:0052428 | modulation of molecular function in other organism(GO:0044359) negative regulation of molecular function in other organism(GO:0044362) negative regulation by host of viral process(GO:0044793) negative regulation of molecular function in other organism involved in symbiotic interaction(GO:0052204) modulation of molecular function in other organism involved in symbiotic interaction(GO:0052205) negative regulation by host of symbiont molecular function(GO:0052405) modification by host of symbiont molecular function(GO:0052428) |
| 0.2 | 0.7 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.2 | 6.1 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.2 | 0.7 | GO:0003162 | atrioventricular node development(GO:0003162) |
| 0.2 | 1.1 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.2 | 1.0 | GO:1900170 | negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.2 | 3.3 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.2 | 0.6 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.2 | 3.4 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.2 | 3.9 | GO:0030903 | notochord development(GO:0030903) |
| 0.2 | 1.8 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.2 | 0.9 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.2 | 1.2 | GO:0015791 | polyol transport(GO:0015791) bile acid secretion(GO:0032782) |
| 0.2 | 0.9 | GO:1904749 | regulation of protein localization to nucleolus(GO:1904749) |
| 0.2 | 2.6 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 1.4 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.1 | 0.7 | GO:0051798 | positive regulation of hair cycle(GO:0042635) positive regulation of hair follicle development(GO:0051798) |
| 0.1 | 0.4 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.1 | 1.7 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.1 | 0.8 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.1 | 1.7 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.1 | 0.5 | GO:0035553 | oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.1 | 0.8 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.1 | 0.5 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.1 | 0.8 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 0.8 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.1 | 1.0 | GO:0061470 | interleukin-21 production(GO:0032625) T follicular helper cell differentiation(GO:0061470) interleukin-21 secretion(GO:0072619) |
| 0.1 | 0.7 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.1 | 0.3 | GO:0002014 | vasoconstriction of artery involved in ischemic response to lowering of systemic arterial blood pressure(GO:0002014) |
| 0.1 | 1.6 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 | 0.2 | GO:0000966 | RNA 5'-end processing(GO:0000966) |
| 0.1 | 0.9 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.1 | 1.3 | GO:0001553 | luteinization(GO:0001553) |
| 0.1 | 0.9 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.1 | 0.4 | GO:0042364 | water-soluble vitamin biosynthetic process(GO:0042364) |
| 0.1 | 5.3 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.1 | 1.7 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.1 | 2.7 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.1 | 0.5 | GO:0021546 | rhombomere development(GO:0021546) |
| 0.1 | 1.2 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.1 | 2.2 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 0.5 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.1 | 0.9 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.1 | 0.8 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.1 | 3.0 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.1 | 8.4 | GO:0030326 | embryonic limb morphogenesis(GO:0030326) embryonic appendage morphogenesis(GO:0035113) |
| 0.1 | 2.6 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.1 | 0.2 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.1 | 1.7 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.1 | 0.2 | GO:0035937 | androgen catabolic process(GO:0006710) estrogen secretion(GO:0035937) estradiol secretion(GO:0035938) regulation of estrogen secretion(GO:2000861) regulation of estradiol secretion(GO:2000864) |
| 0.1 | 3.6 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) |
| 0.1 | 0.6 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 0.6 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 0.6 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.1 | 0.6 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.0 | 0.3 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.1 | GO:0002677 | negative regulation of chronic inflammatory response(GO:0002677) |
| 0.0 | 1.8 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 5.5 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 1.1 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 2.1 | GO:0071774 | response to fibroblast growth factor(GO:0071774) |
| 0.0 | 0.4 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.5 | GO:0006907 | pinocytosis(GO:0006907) |
| 0.0 | 2.5 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.0 | 0.4 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.4 | GO:0014741 | negative regulation of cardiac muscle hypertrophy(GO:0010614) negative regulation of muscle hypertrophy(GO:0014741) |
| 0.0 | 0.7 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 2.1 | GO:0003073 | regulation of systemic arterial blood pressure(GO:0003073) |
| 0.0 | 0.3 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.6 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.0 | 0.3 | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.3 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.0 | 3.2 | GO:0030178 | negative regulation of Wnt signaling pathway(GO:0030178) |
| 0.0 | 0.3 | GO:0035067 | negative regulation of histone acetylation(GO:0035067) |
| 0.0 | 4.4 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.5 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 | 0.3 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 0.8 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.6 | GO:0061028 | establishment of endothelial barrier(GO:0061028) |
| 0.0 | 0.1 | GO:0086023 | adrenergic receptor signaling pathway involved in heart process(GO:0086023) |
| 0.0 | 0.1 | GO:0010719 | negative regulation of epithelial to mesenchymal transition(GO:0010719) |
| 0.0 | 1.4 | GO:0051881 | regulation of mitochondrial membrane potential(GO:0051881) |
| 0.0 | 5.4 | GO:0007059 | chromosome segregation(GO:0007059) |
| 0.0 | 0.6 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.0 | 0.7 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.1 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 1.6 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.0 | 0.1 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 7.3 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.8 | 2.3 | GO:0034684 | integrin alphav-beta5 complex(GO:0034684) |
| 0.7 | 4.1 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.4 | 1.2 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.4 | 1.9 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.3 | 6.8 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.2 | 3.9 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.2 | 0.9 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.2 | 1.0 | GO:0031523 | Myb complex(GO:0031523) |
| 0.2 | 1.6 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.2 | 1.7 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.1 | 5.5 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 0.9 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.1 | 0.7 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.1 | 1.9 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 0.7 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 0.8 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 1.1 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.1 | 3.4 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 0.6 | GO:0061689 | paranodal junction(GO:0033010) tricellular tight junction(GO:0061689) |
| 0.1 | 0.6 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.1 | 4.9 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 0.7 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.1 | 22.0 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 2.3 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 2.7 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 0.3 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 2.6 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 0.8 | GO:0032797 | SMN complex(GO:0032797) |
| 0.1 | 0.9 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.1 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 2.7 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 1.7 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 2.6 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.3 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 1.8 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.7 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 0.9 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 1.7 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.4 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 3.0 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.0 | 0.2 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.8 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 1.1 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 1.8 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.6 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 1.2 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.0 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.5 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 4.9 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 1.4 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.1 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 75.3 | GO:0005634 | nucleus(GO:0005634) |
| 0.0 | 2.6 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 13.1 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.9 | 10.3 | GO:0019531 | secondary active sulfate transmembrane transporter activity(GO:0008271) oxalate transmembrane transporter activity(GO:0019531) |
| 0.9 | 7.5 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.9 | 3.6 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.9 | 4.4 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.8 | 2.5 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.7 | 6.5 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.7 | 4.6 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.6 | 7.3 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.6 | 1.7 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.6 | 7.2 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.5 | 2.5 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.4 | 2.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.3 | 1.4 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.3 | 1.2 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.3 | 0.9 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.3 | 5.1 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.3 | 1.9 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.3 | 1.0 | GO:0038049 | glucocorticoid receptor activity(GO:0004883) transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.3 | 1.8 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.2 | 5.5 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.2 | 8.1 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.2 | 1.7 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.2 | 1.9 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.2 | 2.5 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.2 | 1.2 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.2 | 1.3 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.2 | 1.3 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.2 | 1.4 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.2 | 0.9 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.2 | 3.1 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.2 | 2.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.2 | 1.1 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 3.1 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 3.2 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 0.5 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.1 | 1.7 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 1.1 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.1 | 3.0 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 1.9 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 0.8 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.1 | 10.2 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.1 | 3.4 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 12.6 | GO:0003774 | motor activity(GO:0003774) |
| 0.1 | 1.0 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.1 | 0.3 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.1 | 0.3 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.1 | 0.9 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 2.0 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 1.3 | GO:0000987 | core promoter proximal region sequence-specific DNA binding(GO:0000987) |
| 0.1 | 0.7 | GO:0048185 | activin binding(GO:0048185) |
| 0.1 | 0.4 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 2.1 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 0.4 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.1 | 0.9 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 33.3 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.1 | 0.6 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 0.4 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 1.0 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 0.9 | GO:0015197 | peptide transporter activity(GO:0015197) |
| 0.1 | 0.7 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 0.9 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.1 | 0.4 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 1.9 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 35.7 | GO:0043565 | sequence-specific DNA binding(GO:0043565) |
| 0.0 | 0.4 | GO:0039706 | co-receptor binding(GO:0039706) BMP receptor binding(GO:0070700) |
| 0.0 | 0.8 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 1.2 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.3 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 1.0 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 1.7 | GO:0015144 | carbohydrate transmembrane transporter activity(GO:0015144) carbohydrate transporter activity(GO:1901476) |
| 0.0 | 0.6 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.6 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.1 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.0 | 0.2 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.8 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 0.7 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.3 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 1.3 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.2 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.0 | 0.4 | GO:0001191 | transcriptional repressor activity, RNA polymerase II transcription factor binding(GO:0001191) |
| 0.0 | 0.6 | GO:0043566 | structure-specific DNA binding(GO:0043566) |
| 0.0 | 1.1 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.1 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.9 | GO:0032934 | cholesterol binding(GO:0015485) sterol binding(GO:0032934) |
| 0.0 | 1.8 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.8 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.2 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.0 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 2.0 | GO:0000287 | magnesium ion binding(GO:0000287) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 16.8 | PID_P38_MK2_PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.2 | 18.7 | PID_SMAD2_3NUCLEAR_PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.1 | 2.3 | PID_INTEGRIN5_PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 5.6 | PID_CDC42_REG_PATHWAY | Regulation of CDC42 activity |
| 0.1 | 5.5 | ST_WNT_BETA_CATENIN_PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 1.1 | SA_REG_CASCADE_OF_CYCLIN_EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 1.1 | PID_AR_NONGENOMIC_PATHWAY | Nongenotropic Androgen signaling |
| 0.1 | 3.6 | PID_WNT_NONCANONICAL_PATHWAY | Noncanonical Wnt signaling pathway |
| 0.1 | 6.2 | PID_BETA_CATENIN_NUC_PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 3.3 | PID_NETRIN_PATHWAY | Netrin-mediated signaling events |
| 0.1 | 3.1 | PID_BMP_PATHWAY | BMP receptor signaling |
| 0.1 | 1.8 | PID_EPHRINB_REV_PATHWAY | Ephrin B reverse signaling |
| 0.1 | 1.4 | PID_S1P_S1P3_PATHWAY | S1P3 pathway |
| 0.1 | 6.1 | PID_MYC_ACTIV_PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 1.4 | ST_MYOCYTE_AD_PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 0.8 | PID_SYNDECAN_3_PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 1.1 | PID_HEDGEHOG_2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.6 | NABA_BASEMENT_MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 7.7 | NABA_SECRETED_FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 2.0 | PID_INTEGRIN1_PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 1.7 | PID_ECADHERIN_STABILIZATION_PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 2.4 | PID_AR_PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 4.4 | NABA_ECM_REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.2 | PID_MYC_PATHWAY | C-MYC pathway |
| 0.0 | 0.6 | PID_DNA_PK_PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.5 | PID_MAPK_TRK_PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.4 | PID_IL2_STAT5_PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 1.0 | ST_ERK1_ERK2_MAPK_PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 1.3 | PID_PLK1_PATHWAY | PLK1 signaling events |
| 0.0 | 1.0 | PID_FANCONI_PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.8 | PID_RAC1_PATHWAY | RAC1 signaling pathway |
| 0.0 | 2.0 | NABA_ECM_GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.7 | PID_NOTCH_PATHWAY | Notch signaling pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 8.6 | REACTOME_FGFR1_LIGAND_BINDING_AND_ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.4 | 1.1 | REACTOME_HORMONE_LIGAND_BINDING_RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.2 | 12.0 | REACTOME_AMINO_ACID_AND_OLIGOPEPTIDE_SLC_TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.2 | 1.9 | REACTOME_GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.2 | 4.6 | REACTOME_GABA_SYNTHESIS_RELEASE_REUPTAKE_AND_DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.2 | 9.7 | REACTOME_TRANSPORT_OF_INORGANIC_CATIONS_ANIONS_AND_AMINO_ACIDS_OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.2 | 1.2 | REACTOME_PASSIVE_TRANSPORT_BY_AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 2.3 | REACTOME_PECAM1_INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.1 | 2.1 | REACTOME_IONOTROPIC_ACTIVITY_OF_KAINATE_RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 1.0 | REACTOME_ERKS_ARE_INACTIVATED | Genes involved in ERKs are inactivated |
| 0.1 | 4.9 | REACTOME_NETRIN1_SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 3.1 | REACTOME_DOWNREGULATION_OF_SMAD2_3_SMAD4_TRANSCRIPTIONAL_ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.1 | 9.1 | REACTOME_MITOTIC_PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 1.9 | REACTOME_SIGNALING_BY_BMP | Genes involved in Signaling by BMP |
| 0.1 | 1.1 | REACTOME_CYCLIN_A_B1_ASSOCIATED_EVENTS_DURING_G2_M_TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 1.7 | REACTOME_SULFUR_AMINO_ACID_METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 3.5 | REACTOME_RNA_POL_I_PROMOTER_OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.1 | 1.4 | REACTOME_MITOCHONDRIAL_TRNA_AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 0.8 | REACTOME_REGULATION_OF_KIT_SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.1 | 0.5 | REACTOME_P2Y_RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 0.9 | REACTOME_FORMATION_OF_ATP_BY_CHEMIOSMOTIC_COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 0.9 | REACTOME_RNA_POL_I_TRANSCRIPTION_TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 1.7 | REACTOME_G1_PHASE | Genes involved in G1 Phase |
| 0.0 | 1.8 | REACTOME_HS_GAG_BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.7 | REACTOME_NOTCH_HLH_TRANSCRIPTION_PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 1.2 | REACTOME_ABC_FAMILY_PROTEINS_MEDIATED_TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 1.2 | REACTOME_MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.9 | REACTOME_STRIATED_MUSCLE_CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.5 | REACTOME_APOPTOSIS_INDUCED_DNA_FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.9 | REACTOME_BIOSYNTHESIS_OF_THE_N_GLYCAN_PRECURSOR_DOLICHOL_LIPID_LINKED_OLIGOSACCHARIDE_LLO_AND_TRANSFER_TO_A_NASCENT_PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 1.0 | REACTOME_BMAL1_CLOCK_NPAS2_ACTIVATES_CIRCADIAN_EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 1.5 | REACTOME_COLLAGEN_FORMATION | Genes involved in Collagen formation |
| 0.0 | 1.5 | REACTOME_CLASS_B_2_SECRETIN_FAMILY_RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.3 | REACTOME_KINESINS | Genes involved in Kinesins |
| 0.0 | 0.4 | REACTOME_RNA_POL_II_TRANSCRIPTION_PRE_INITIATION_AND_PROMOTER_OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.1 | REACTOME_ACYL_CHAIN_REMODELLING_OF_PG | Genes involved in Acyl chain remodelling of PG |