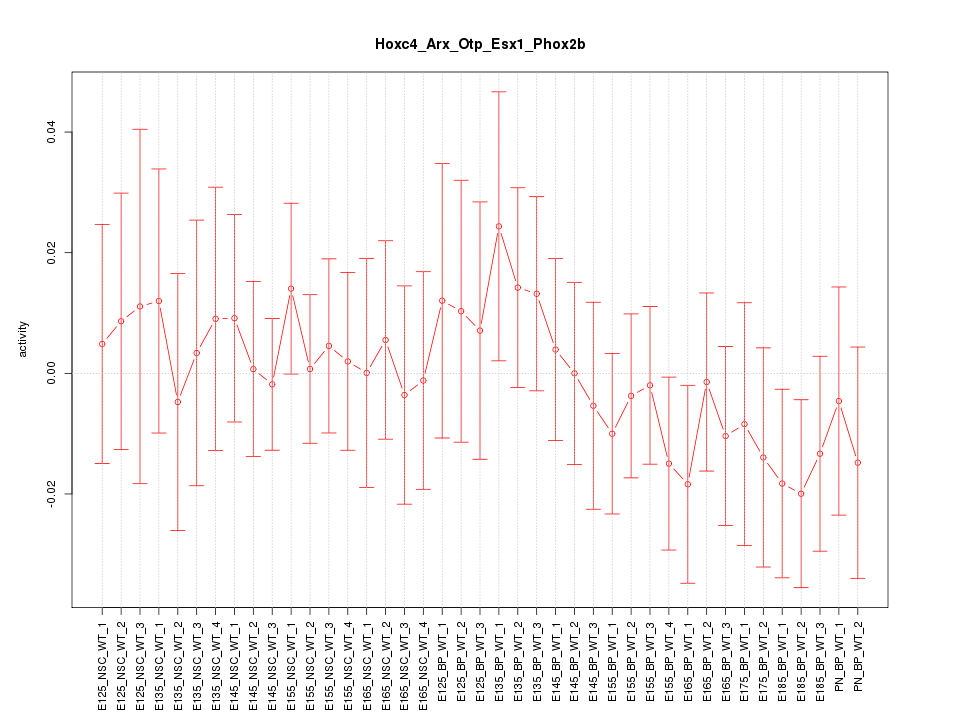
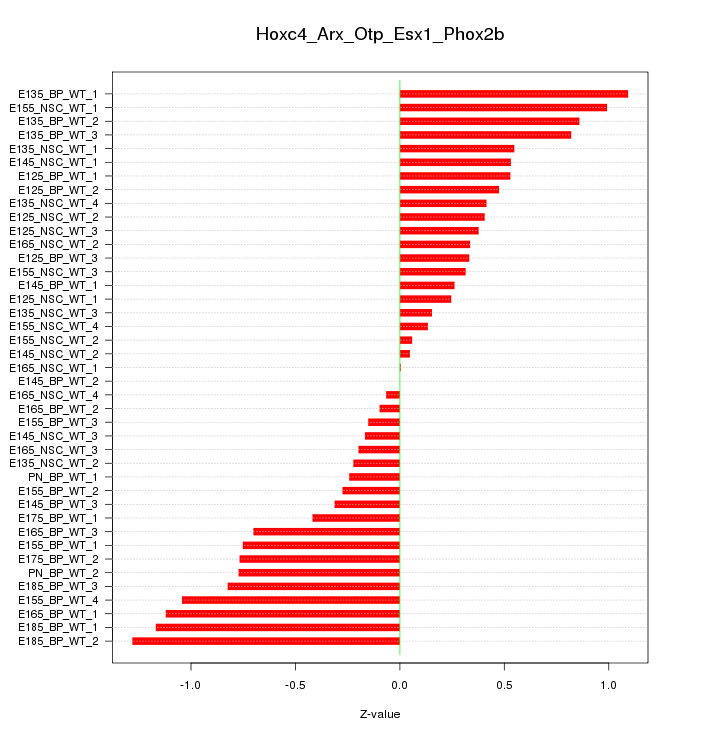
Motif ID: Hoxc4_Arx_Otp_Esx1_Phox2b
Z-value: 0.595
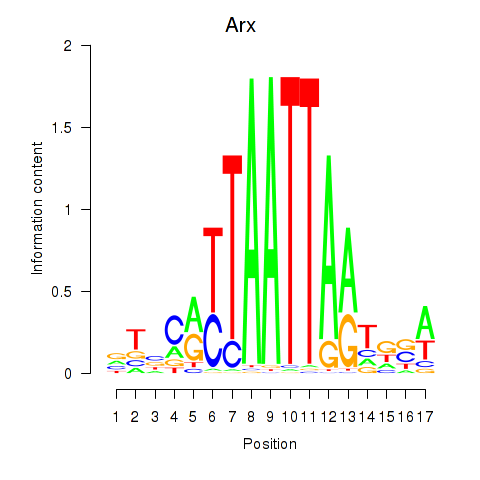
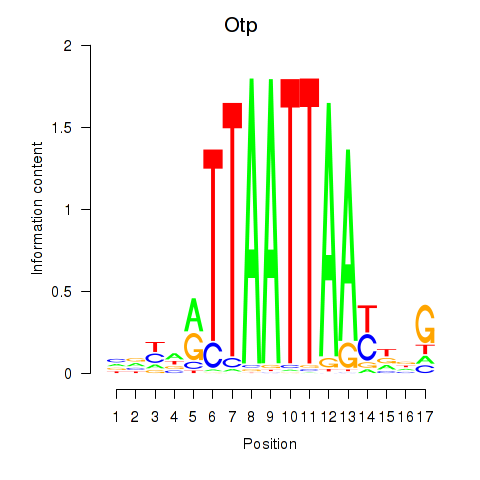
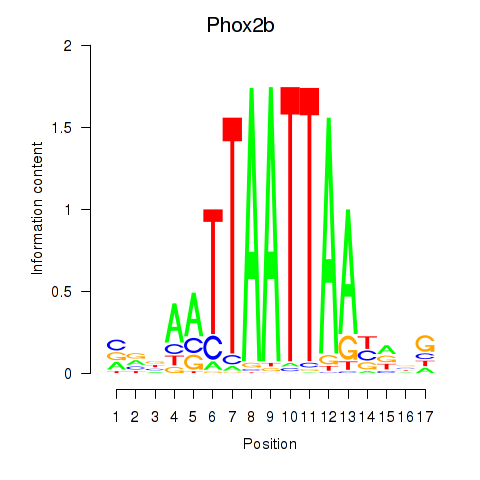
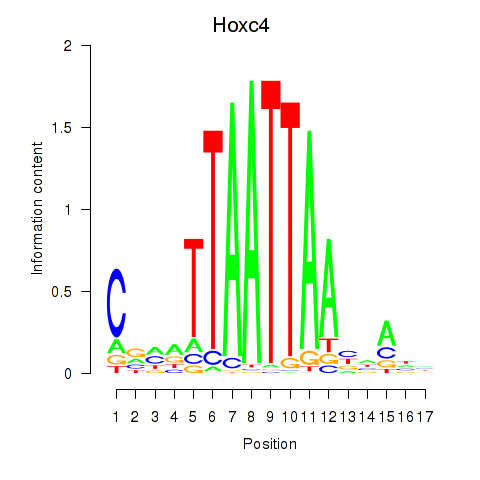
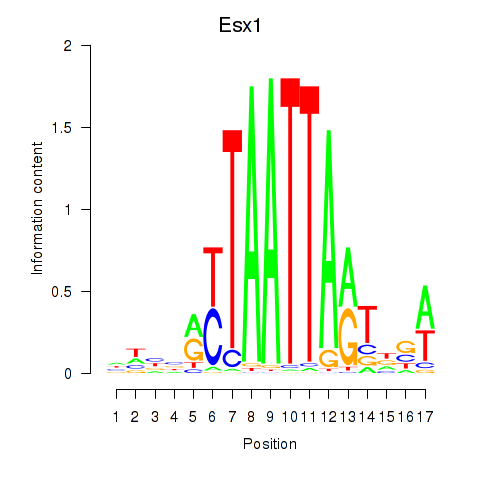
Transcription factors associated with Hoxc4_Arx_Otp_Esx1_Phox2b:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Arx | ENSMUSG00000035277.9 | Arx |
| Esx1 | ENSMUSG00000023443.7 | Esx1 |
| Hoxc4 | ENSMUSG00000075394.3 | Hoxc4 |
| Otp | ENSMUSG00000021685.10 | Otp |
| Phox2b | ENSMUSG00000012520.8 | Phox2b |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Otp | mm10_v2_chr13_+_94875600_94875611 | 0.41 | 7.5e-03 | Click! |
| Arx | mm10_v2_chrX_+_93286499_93286522 | 0.35 | 2.7e-02 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.4 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.8 | 2.4 | GO:0031660 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) |
| 0.5 | 2.1 | GO:0010749 | regulation of nitric oxide mediated signal transduction(GO:0010749) |
| 0.5 | 3.6 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.4 | 1.2 | GO:0035934 | corticosterone secretion(GO:0035934) regulation of corticosterone secretion(GO:2000852) |
| 0.3 | 0.9 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.2 | 4.5 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.2 | 1.5 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.2 | 0.6 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.2 | 4.0 | GO:0007530 | sex determination(GO:0007530) |
| 0.2 | 0.9 | GO:0072235 | distal convoluted tubule development(GO:0072025) metanephric distal convoluted tubule development(GO:0072221) metanephric distal tubule development(GO:0072235) |
| 0.2 | 0.5 | GO:0046098 | regulation of primitive erythrocyte differentiation(GO:0010725) guanine metabolic process(GO:0046098) |
| 0.2 | 1.0 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.2 | 0.5 | GO:0060489 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.2 | 0.6 | GO:0072592 | regulation of integrin biosynthetic process(GO:0045113) oxygen metabolic process(GO:0072592) |
| 0.2 | 1.2 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.1 | 0.4 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.1 | 0.8 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.1 | 1.3 | GO:0036376 | sodium ion export from cell(GO:0036376) |
| 0.1 | 2.7 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.1 | 0.7 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.1 | 2.3 | GO:0071803 | keratinocyte development(GO:0003334) positive regulation of podosome assembly(GO:0071803) |
| 0.1 | 0.3 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.1 | 0.5 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.1 | 4.1 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.1 | 0.4 | GO:0046950 | cellular ketone body metabolic process(GO:0046950) |
| 0.1 | 0.6 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.1 | 0.9 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 0.5 | GO:0098535 | positive regulation of centriole replication(GO:0046601) de novo centriole assembly(GO:0098535) |
| 0.1 | 0.5 | GO:0060923 | cardiac muscle cell fate commitment(GO:0060923) |
| 0.1 | 0.4 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.1 | 0.5 | GO:0072615 | interleukin-17 secretion(GO:0072615) |
| 0.1 | 0.5 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) protein oxidation(GO:0018158) |
| 0.1 | 2.2 | GO:0021591 | ventricular system development(GO:0021591) |
| 0.1 | 0.6 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.1 | 1.2 | GO:0002183 | cytoplasmic translational initiation(GO:0002183) |
| 0.1 | 0.2 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.1 | 0.2 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.4 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.0 | 0.8 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.7 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 2.1 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 1.6 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) long-term memory(GO:0007616) |
| 0.0 | 0.3 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.0 | 0.7 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.0 | 4.9 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.0 | 1.0 | GO:0006907 | pinocytosis(GO:0006907) |
| 0.0 | 0.1 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.2 | GO:0036337 | Fas signaling pathway(GO:0036337) |
| 0.0 | 0.4 | GO:1902916 | positive regulation of protein polyubiquitination(GO:1902916) |
| 0.0 | 0.2 | GO:1900116 | sequestering of extracellular ligand from receptor(GO:0035581) determination of dorsal/ventral asymmetry(GO:0048262) determination of dorsal identity(GO:0048263) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.0 | GO:0035482 | gastric motility(GO:0035482) gastric emptying(GO:0035483) |
| 0.0 | 0.1 | GO:0035553 | oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.0 | 0.6 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.0 | 0.6 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 0.3 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.0 | 0.2 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.0 | 0.2 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.1 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 | 0.3 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.7 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.1 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.0 | 1.9 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.1 | GO:1901509 | positive regulation of cAMP-mediated signaling(GO:0043950) regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.0 | 0.2 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.2 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.1 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 0.6 | GO:1901381 | positive regulation of potassium ion transmembrane transport(GO:1901381) |
| 0.0 | 0.2 | GO:0060837 | blood vessel endothelial cell differentiation(GO:0060837) |
| 0.0 | 0.1 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.0 | 1.5 | GO:2000177 | regulation of neural precursor cell proliferation(GO:2000177) |
| 0.0 | 0.3 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.6 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.0 | 0.1 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.0 | 0.3 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.0 | 0.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 4.0 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 0.5 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 0.6 | GO:0002066 | columnar/cuboidal epithelial cell development(GO:0002066) |
| 0.0 | 1.2 | GO:0031497 | chromatin assembly(GO:0031497) |
| 0.0 | 0.1 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.7 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.6 | 2.4 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.2 | 1.3 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.2 | 0.6 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.1 | 1.0 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.1 | 1.2 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.1 | 0.5 | GO:0060187 | cell pole(GO:0060187) |
| 0.1 | 0.7 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.1 | 0.6 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 1.6 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 0.5 | GO:0098536 | deuterosome(GO:0098536) |
| 0.1 | 1.0 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.1 | 0.5 | GO:0097413 | Lewy body(GO:0097413) |
| 0.1 | 0.2 | GO:0031417 | NatC complex(GO:0031417) |
| 0.1 | 0.6 | GO:0034709 | methylosome(GO:0034709) |
| 0.1 | 2.3 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.4 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 1.8 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 2.2 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.2 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 0.2 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.0 | 4.5 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.0 | 0.5 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.9 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 0.8 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.4 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 1.4 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.4 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.3 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.2 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.1 | GO:0030689 | Noc complex(GO:0030689) |
| 0.0 | 0.2 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.2 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.7 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 0.2 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.1 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.2 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.4 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.6 | 2.4 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.5 | 1.9 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.4 | 1.2 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.4 | 1.2 | GO:0031779 | melanocortin receptor binding(GO:0031779) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.3 | 3.6 | GO:0019531 | secondary active sulfate transmembrane transporter activity(GO:0008271) oxalate transmembrane transporter activity(GO:0019531) |
| 0.3 | 1.6 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.2 | 0.6 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.1 | 1.0 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 0.7 | GO:0034604 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.1 | 0.5 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.1 | 1.3 | GO:0008556 | sodium:potassium-exchanging ATPase activity(GO:0005391) potassium-transporting ATPase activity(GO:0008556) |
| 0.1 | 0.8 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.1 | 4.1 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.1 | 0.6 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.1 | 0.4 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.1 | 0.5 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.1 | 3.1 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 0.8 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.1 | 0.3 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.1 | 0.9 | GO:0031957 | fatty acid transporter activity(GO:0015245) very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 0.4 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.0 | 1.0 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 9.0 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.3 | GO:0008297 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) |
| 0.0 | 0.3 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 1.6 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.5 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.0 | 0.6 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 2.1 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.2 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.1 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.0 | 0.2 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.3 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.0 | 1.3 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.4 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.2 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.1 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.0 | 0.4 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) |
| 0.0 | 0.6 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.2 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.2 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.5 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.2 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.2 | GO:0045125 | bioactive lipid receptor activity(GO:0045125) |
| 0.0 | 4.4 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 0.3 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.7 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.5 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.1 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.2 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.1 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 0.2 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.0 | GO:0004816 | asparagine-tRNA ligase activity(GO:0004816) |
| 0.0 | 0.4 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.1 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.0 | 0.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.9 | GO:0019842 | vitamin binding(GO:0019842) |
| 0.0 | 0.0 | GO:0052740 | phosphatidylserine 1-acylhydrolase activity(GO:0052739) 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.0 | 0.2 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 1.2 | GO:0016779 | nucleotidyltransferase activity(GO:0016779) |
| 0.0 | 0.6 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | SA_REG_CASCADE_OF_CYCLIN_EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 1.6 | PID_LPA4_PATHWAY | LPA4-mediated signaling events |
| 0.1 | 2.7 | PID_FOXM1_PATHWAY | FOXM1 transcription factor network |
| 0.1 | 1.2 | PID_SYNDECAN_3_PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.5 | PID_ERB_GENOMIC_PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 3.3 | PID_BETA_CATENIN_NUC_PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 1.1 | PID_AURORA_B_PATHWAY | Aurora B signaling |
| 0.0 | 2.0 | PID_AR_PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.7 | NABA_BASEMENT_MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.2 | SA_PROGRAMMED_CELL_DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.8 | PID_CASPASE_PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.4 | PID_NCADHERIN_PATHWAY | N-cadherin signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.7 | REACTOME_REGULATION_OF_INSULIN_SECRETION_BY_ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.3 | 1.5 | REACTOME_CDC6_ASSOCIATION_WITH_THE_ORC_ORIGIN_COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.2 | 2.4 | REACTOME_G2_M_DNA_DAMAGE_CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.2 | 1.2 | REACTOME_ANDROGEN_BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.2 | 1.9 | REACTOME_TERMINATION_OF_O_GLYCAN_BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 1.6 | REACTOME_ADENYLATE_CYCLASE_ACTIVATING_PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 0.9 | REACTOME_SYNTHESIS_OF_BILE_ACIDS_AND_BILE_SALTS_VIA_24_HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.1 | 0.7 | REACTOME_REGULATION_OF_PYRUVATE_DEHYDROGENASE_PDH_COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.5 | REACTOME_PURINE_CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 1.4 | REACTOME_ADHERENS_JUNCTIONS_INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 1.3 | REACTOME_ION_TRANSPORT_BY_P_TYPE_ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 1.8 | REACTOME_DEPOSITION_OF_NEW_CENPA_CONTAINING_NUCLEOSOMES_AT_THE_CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.6 | REACTOME_RETROGRADE_NEUROTROPHIN_SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.2 | REACTOME_PRESYNAPTIC_NICOTINIC_ACETYLCHOLINE_RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.5 | REACTOME_MITOCHONDRIAL_TRNA_AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 2.3 | REACTOME_TRANSPORT_OF_INORGANIC_CATIONS_ANIONS_AND_AMINO_ACIDS_OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.3 | REACTOME_POST_CHAPERONIN_TUBULIN_FOLDING_PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 1.0 | REACTOME_GOLGI_ASSOCIATED_VESICLE_BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 1.1 | REACTOME_CLASS_B_2_SECRETIN_FAMILY_RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.2 | REACTOME_UNWINDING_OF_DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.7 | REACTOME_RESPIRATORY_ELECTRON_TRANSPORT | Genes involved in Respiratory electron transport |