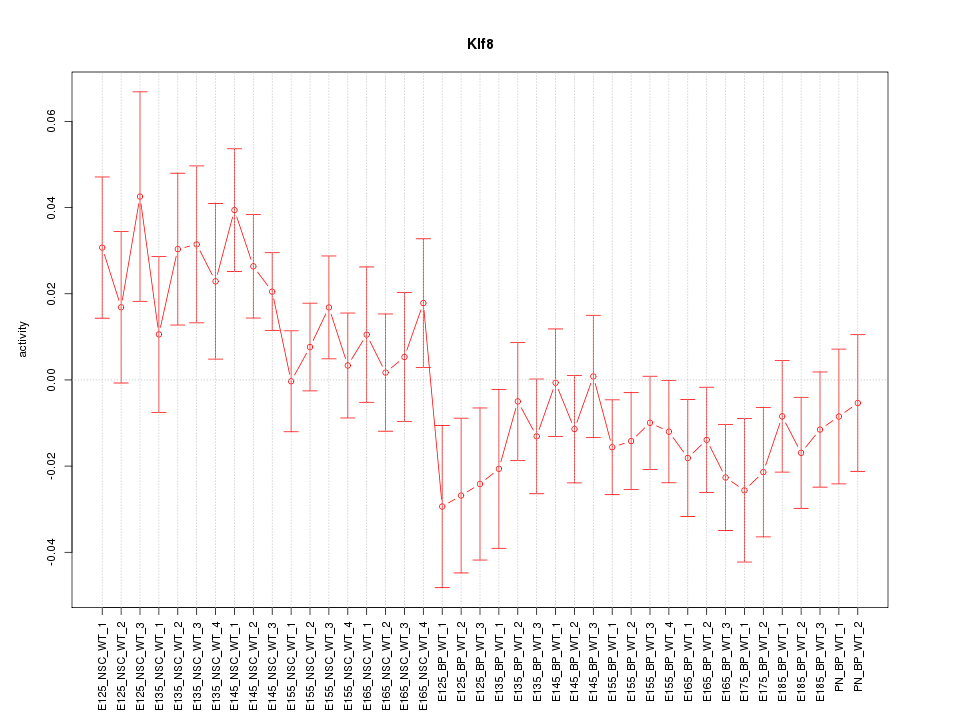
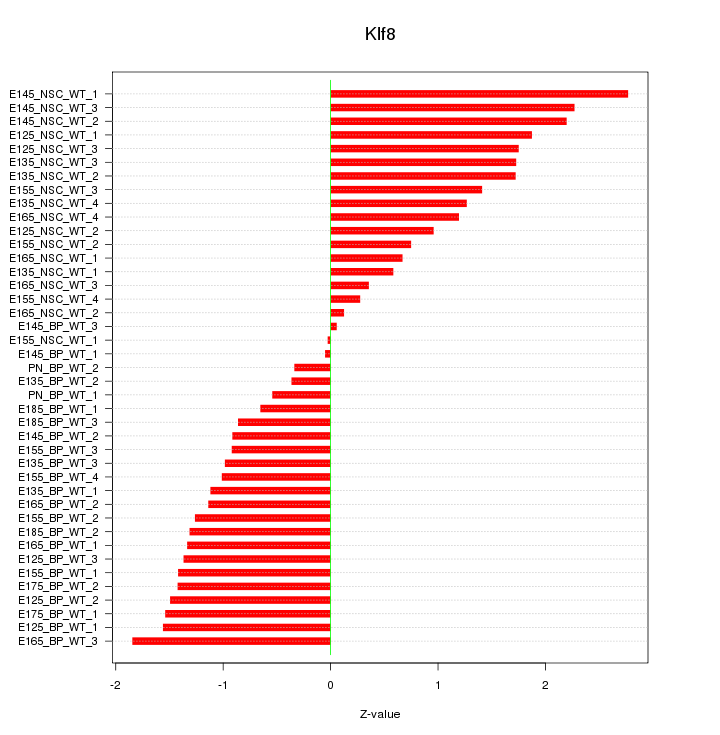
Motif ID: Klf8
Z-value: 1.279

Transcription factors associated with Klf8:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Klf8 | ENSMUSG00000041649.7 | Klf8 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Klf8 | mm10_v2_chrX_+_153237748_153237748 | -0.32 | 3.8e-02 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 12.7 | GO:0046022 | regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 2.8 | 8.5 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 2.5 | 10.2 | GO:0010986 | positive regulation of lipoprotein particle clearance(GO:0010986) |
| 2.2 | 6.5 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 1.6 | 9.6 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 1.6 | 6.3 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 1.6 | 9.4 | GO:0060242 | contact inhibition(GO:0060242) |
| 1.5 | 5.9 | GO:0060745 | mammary gland branching involved in pregnancy(GO:0060745) |
| 1.4 | 8.6 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 1.3 | 4.0 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 1.2 | 3.6 | GO:0060598 | dichotomous subdivision of terminal units involved in mammary gland duct morphogenesis(GO:0060598) |
| 1.1 | 3.2 | GO:0036166 | phenotypic switching(GO:0036166) |
| 1.0 | 3.1 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 1.0 | 9.6 | GO:0042473 | outer ear morphogenesis(GO:0042473) |
| 1.0 | 6.7 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.9 | 14.7 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.9 | 2.6 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.9 | 2.6 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.8 | 2.4 | GO:0010641 | positive regulation of platelet-derived growth factor receptor signaling pathway(GO:0010641) |
| 0.8 | 3.9 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.7 | 2.2 | GO:0036388 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) DNA replication preinitiation complex assembly(GO:0071163) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.7 | 3.7 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.7 | 3.7 | GO:0046668 | regulation of retinal cell programmed cell death(GO:0046668) |
| 0.7 | 0.7 | GO:1902460 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.7 | 4.7 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.7 | 4.6 | GO:0000320 | re-entry into mitotic cell cycle(GO:0000320) |
| 0.7 | 3.3 | GO:0018002 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.6 | 3.8 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.6 | 2.5 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.6 | 5.0 | GO:0072615 | interleukin-17 secretion(GO:0072615) |
| 0.6 | 1.8 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.6 | 3.6 | GO:0002317 | plasma cell differentiation(GO:0002317) positive regulation of viral entry into host cell(GO:0046598) |
| 0.6 | 2.9 | GO:0051309 | female meiosis chromosome separation(GO:0051309) |
| 0.6 | 2.3 | GO:0015744 | succinate transport(GO:0015744) |
| 0.4 | 0.8 | GO:1903224 | regulation of endodermal cell fate specification(GO:0042663) regulation of endodermal cell differentiation(GO:1903224) |
| 0.4 | 2.9 | GO:2001185 | regulation of CD8-positive, alpha-beta T cell activation(GO:2001185) |
| 0.4 | 16.1 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.3 | 1.4 | GO:0007056 | spindle assembly involved in female meiosis(GO:0007056) |
| 0.3 | 3.7 | GO:1900121 | negative regulation of receptor binding(GO:1900121) |
| 0.3 | 2.3 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.3 | 1.3 | GO:0060265 | positive regulation of respiratory burst involved in inflammatory response(GO:0060265) |
| 0.3 | 3.3 | GO:0042487 | regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.3 | 4.1 | GO:0099628 | receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 0.3 | 7.0 | GO:0043486 | histone exchange(GO:0043486) |
| 0.3 | 0.9 | GO:0002408 | myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.3 | 1.5 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.3 | 0.9 | GO:0061181 | regulation of chondrocyte development(GO:0061181) |
| 0.3 | 1.1 | GO:1904706 | regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010578) positive regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010579) negative regulation of vascular smooth muscle cell proliferation(GO:1904706) positive regulation of vascular smooth muscle cell differentiation(GO:1905065) |
| 0.3 | 1.4 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.3 | 1.1 | GO:0035553 | oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.3 | 1.6 | GO:0032439 | endosome localization(GO:0032439) |
| 0.3 | 0.8 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.3 | 1.3 | GO:0019346 | homoserine metabolic process(GO:0009092) transsulfuration(GO:0019346) |
| 0.3 | 1.3 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.3 | 1.8 | GO:1902965 | cellular response to testosterone stimulus(GO:0071394) regulation of protein localization to early endosome(GO:1902965) positive regulation of protein localization to early endosome(GO:1902966) |
| 0.3 | 1.3 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.2 | 1.5 | GO:1903275 | positive regulation of sodium ion export(GO:1903275) positive regulation of sodium ion export from cell(GO:1903278) regulation of inorganic anion transmembrane transport(GO:1903795) |
| 0.2 | 2.2 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.2 | 1.7 | GO:0097421 | liver regeneration(GO:0097421) |
| 0.2 | 2.4 | GO:0048385 | regulation of retinoic acid receptor signaling pathway(GO:0048385) |
| 0.2 | 2.1 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.2 | 0.7 | GO:0010725 | regulation of primitive erythrocyte differentiation(GO:0010725) |
| 0.2 | 1.5 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.2 | 1.1 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.2 | 0.8 | GO:0060578 | extraocular skeletal muscle development(GO:0002074) subthalamic nucleus development(GO:0021763) left lung development(GO:0060459) left lung morphogenesis(GO:0060460) pulmonary vein morphogenesis(GO:0060577) superior vena cava morphogenesis(GO:0060578) |
| 0.2 | 7.3 | GO:0009299 | mRNA transcription(GO:0009299) |
| 0.2 | 1.0 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.2 | 1.5 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.2 | 1.1 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.2 | 4.1 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.2 | 0.8 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.2 | 5.7 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.2 | 1.0 | GO:0097646 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.2 | 2.4 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.2 | 0.6 | GO:0006447 | regulation of translational initiation by iron(GO:0006447) positive regulation of translational initiation by iron(GO:0045994) |
| 0.1 | 1.8 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.1 | 1.0 | GO:0031086 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.1 | 2.0 | GO:0071670 | smooth muscle cell chemotaxis(GO:0071670) |
| 0.1 | 2.2 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.1 | 2.7 | GO:0042711 | maternal behavior(GO:0042711) |
| 0.1 | 1.9 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.1 | 0.4 | GO:1902949 | positive regulation of tau-protein kinase activity(GO:1902949) |
| 0.1 | 2.1 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.1 | 0.4 | GO:0060489 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.1 | 4.4 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 1.9 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.1 | 1.4 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.1 | 0.4 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.1 | 0.7 | GO:0031508 | chromatin remodeling at centromere(GO:0031055) pericentric heterochromatin assembly(GO:0031508) |
| 0.1 | 1.4 | GO:0060033 | anatomical structure regression(GO:0060033) |
| 0.1 | 2.3 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.1 | 0.5 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.1 | 0.6 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.1 | 0.2 | GO:0003256 | regulation of transcription from RNA polymerase II promoter involved in myocardial precursor cell differentiation(GO:0003256) |
| 0.1 | 1.4 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.1 | 1.2 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.1 | 0.7 | GO:0060019 | radial glial cell differentiation(GO:0060019) |
| 0.1 | 0.2 | GO:0019401 | alditol biosynthetic process(GO:0019401) |
| 0.1 | 2.1 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.1 | 0.4 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) establishment of Sertoli cell barrier(GO:0097368) |
| 0.1 | 2.2 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.1 | 0.8 | GO:0033008 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.1 | 0.2 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.1 | 1.0 | GO:0002467 | germinal center formation(GO:0002467) |
| 0.1 | 0.7 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.1 | 0.9 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 0.4 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.1 | 0.6 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 0.1 | 0.2 | GO:0014005 | microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.1 | 0.5 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.1 | 0.8 | GO:0034982 | mitochondrial protein processing(GO:0034982) |
| 0.0 | 0.6 | GO:0042182 | ketone catabolic process(GO:0042182) |
| 0.0 | 0.6 | GO:0070633 | transepithelial transport(GO:0070633) |
| 0.0 | 1.0 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 1.5 | GO:1903077 | negative regulation of protein localization to plasma membrane(GO:1903077) negative regulation of protein localization to cell periphery(GO:1904376) |
| 0.0 | 1.2 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.2 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.8 | GO:0044381 | glucose import in response to insulin stimulus(GO:0044381) |
| 0.0 | 0.2 | GO:0000019 | regulation of mitotic recombination(GO:0000019) telomeric 3' overhang formation(GO:0031860) |
| 0.0 | 0.4 | GO:0046697 | decidualization(GO:0046697) |
| 0.0 | 0.8 | GO:0030279 | negative regulation of ossification(GO:0030279) |
| 0.0 | 0.8 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 0.2 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 2.6 | GO:0050709 | negative regulation of protein secretion(GO:0050709) |
| 0.0 | 0.1 | GO:0060903 | positive regulation of meiosis I(GO:0060903) |
| 0.0 | 2.2 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.2 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.5 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 1.0 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 1.2 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.2 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 0.3 | GO:0045780 | positive regulation of bone resorption(GO:0045780) positive regulation of bone remodeling(GO:0046852) |
| 0.0 | 1.7 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 0.2 | GO:0046498 | S-adenosylhomocysteine metabolic process(GO:0046498) |
| 0.0 | 0.7 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.4 | GO:0006073 | glycogen metabolic process(GO:0005977) cellular glucan metabolic process(GO:0006073) glucan metabolic process(GO:0044042) cellular polysaccharide metabolic process(GO:0044264) |
| 0.0 | 0.1 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.0 | 0.5 | GO:0032330 | regulation of chondrocyte differentiation(GO:0032330) |
| 0.0 | 0.4 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.0 | 0.8 | GO:0006096 | glycolytic process(GO:0006096) ATP generation from ADP(GO:0006757) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.4 | 16.1 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 3.1 | 9.4 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 2.6 | 7.8 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 1.4 | 4.1 | GO:0099573 | glutamatergic postsynaptic density(GO:0099573) |
| 1.2 | 7.3 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.9 | 2.6 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.8 | 5.9 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.8 | 3.8 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.7 | 2.2 | GO:0036387 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.7 | 4.0 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.6 | 3.7 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.6 | 1.7 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.5 | 1.5 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.4 | 3.3 | GO:0031415 | NatA complex(GO:0031415) |
| 0.4 | 5.7 | GO:0042555 | MCM complex(GO:0042555) |
| 0.3 | 10.2 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.3 | 2.9 | GO:0000796 | condensin complex(GO:0000796) |
| 0.3 | 0.8 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.2 | 1.5 | GO:0034448 | EGO complex(GO:0034448) |
| 0.2 | 1.4 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.2 | 0.8 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.2 | 1.8 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.2 | 2.0 | GO:0046930 | pore complex(GO:0046930) |
| 0.1 | 1.0 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.1 | 3.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 0.4 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.1 | 1.1 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 0.9 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 0.5 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 0.8 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 0.4 | GO:0060187 | cell pole(GO:0060187) |
| 0.1 | 1.2 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.1 | 1.4 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 0.8 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 6.6 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 0.4 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.1 | 1.1 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 1.1 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.5 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 17.4 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 2.5 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 1.0 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 1.6 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.5 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.8 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.7 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.8 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 0.5 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 1.3 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 1.0 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.2 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 1.9 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 6.0 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.4 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 1.2 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.6 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 1.5 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 1.5 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 0.8 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 1.2 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.1 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 3.9 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 1.5 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.4 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 1.5 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.0 | 5.1 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.6 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.0 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.7 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.5 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) integral component of Golgi membrane(GO:0030173) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.0 | 16.1 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 2.8 | 8.5 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 2.5 | 10.2 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 2.0 | 5.9 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 1.2 | 5.0 | GO:0008142 | oxysterol binding(GO:0008142) |
| 1.1 | 6.5 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.9 | 2.6 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.8 | 2.3 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.7 | 3.7 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.7 | 3.6 | GO:0005534 | galactose binding(GO:0005534) |
| 0.7 | 14.7 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.7 | 3.3 | GO:1990190 | peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.5 | 3.2 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.4 | 12.4 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.4 | 1.7 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.4 | 2.4 | GO:0019841 | retinol binding(GO:0019841) |
| 0.4 | 1.1 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.3 | 4.1 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.3 | 1.0 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 0.3 | 1.0 | GO:0004658 | propionyl-CoA carboxylase activity(GO:0004658) |
| 0.3 | 1.6 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.3 | 7.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.3 | 3.7 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.3 | 1.1 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.3 | 1.3 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.3 | 1.3 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.3 | 4.0 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.2 | 1.7 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.2 | 2.0 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.2 | 0.6 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.2 | 0.8 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.2 | 0.4 | GO:0004921 | interleukin-11 receptor activity(GO:0004921) interleukin-11 binding(GO:0019970) |
| 0.2 | 0.8 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.2 | 1.4 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.2 | 0.6 | GO:0036470 | tyrosine 3-monooxygenase activator activity(GO:0036470) L-dopa decarboxylase activator activity(GO:0036478) |
| 0.2 | 2.6 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.2 | 2.2 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.2 | 2.7 | GO:0051861 | glycolipid binding(GO:0051861) |
| 0.2 | 0.6 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.2 | 1.1 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.1 | 3.8 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 3.1 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 2.2 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.1 | 1.4 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 0.6 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.1 | 0.9 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 2.2 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 4.0 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.1 | 0.4 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.1 | 0.4 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.1 | 1.7 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 8.6 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.1 | 1.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 1.8 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 0.9 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 5.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 1.6 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 2.1 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 0.6 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.1 | 0.8 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 1.5 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.1 | 0.2 | GO:0098973 | structural constituent of synapse(GO:0098918) structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.1 | 0.9 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 0.4 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.1 | 0.2 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.1 | 1.0 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.1 | 0.8 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 5.0 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.1 | 0.2 | GO:0008967 | phosphoglycolate phosphatase activity(GO:0008967) |
| 0.1 | 1.5 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.5 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.2 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 5.1 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 1.3 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.7 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.2 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 0.5 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.8 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 1.6 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 1.4 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 3.1 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.1 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.0 | 0.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.7 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 4.9 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 0.9 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 3.5 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.8 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.0 | 0.1 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.0 | 1.5 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.4 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.3 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.4 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 1.6 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 1.1 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 1.1 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 3.3 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 6.5 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 0.1 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.0 | 1.8 | GO:0000981 | RNA polymerase II transcription factor activity, sequence-specific DNA binding(GO:0000981) |
| 0.0 | 1.4 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 7.2 | GO:0043565 | sequence-specific DNA binding(GO:0043565) |
| 0.0 | 2.9 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.6 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.1 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 3.9 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.0 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.8 | GO:0008238 | exopeptidase activity(GO:0008238) |
| 0.0 | 0.4 | GO:0048365 | Rac GTPase binding(GO:0048365) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 24.7 | PID_DELTA_NP63_PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.4 | 19.8 | PID_FOXM1_PATHWAY | FOXM1 transcription factor network |
| 0.2 | 2.0 | SA_FAS_SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.2 | 3.9 | PID_INTEGRIN5_PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.2 | 4.1 | ST_WNT_CA2_CYCLIC_GMP_PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.2 | 15.8 | PID_SMAD2_3NUCLEAR_PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.2 | 9.1 | PID_ARF6_TRAFFICKING_PATHWAY | Arf6 trafficking events |
| 0.1 | 6.5 | PID_E2F_PATHWAY | E2F transcription factor network |
| 0.1 | 3.2 | PID_TCR_CALCIUM_PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 1.5 | PID_INSULIN_PATHWAY | Insulin Pathway |
| 0.1 | 3.7 | PID_BMP_PATHWAY | BMP receptor signaling |
| 0.1 | 3.0 | PID_TAP63_PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 3.8 | PID_AR_TF_PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 1.9 | NABA_BASEMENT_MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 1.8 | ST_GAQ_PATHWAY | G alpha q Pathway |
| 0.1 | 0.9 | PID_BETA_CATENIN_DEG_PATHWAY | Degradation of beta catenin |
| 0.0 | 5.6 | NABA_ECM_AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.9 | PID_ILK_PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 1.2 | PID_WNT_SIGNALING_PATHWAY | Wnt signaling network |
| 0.0 | 2.5 | PID_P53_REGULATION_PATHWAY | p53 pathway |
| 0.0 | 1.1 | PID_AURORA_A_PATHWAY | Aurora A signaling |
| 0.0 | 2.2 | PID_REG_GR_PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 2.9 | PID_RHOA_REG_PATHWAY | Regulation of RhoA activity |
| 0.0 | 2.0 | PID_HDAC_CLASSI_PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.6 | PID_ALPHA_SYNUCLEIN_PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.7 | PID_HNF3B_PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.0 | PID_PLK1_PATHWAY | PLK1 signaling events |
| 0.0 | 0.9 | PID_ER_NONGENOMIC_PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 0.4 | PID_HES_HEY_PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 1.5 | NABA_ECM_GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 2.1 | NABA_SECRETED_FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.4 | ST_ERK1_ERK2_MAPK_PATHWAY | ERK1/ERK2 MAPK Pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 8.0 | REACTOME_UNWINDING_OF_DNA | Genes involved in Unwinding of DNA |
| 0.5 | 16.8 | REACTOME_DOWNREGULATION_OF_SMAD2_3_SMAD4_TRANSCRIPTIONAL_ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.4 | 4.7 | REACTOME_FGFR1_LIGAND_BINDING_AND_ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.4 | 13.0 | REACTOME_NUCLEAR_SIGNALING_BY_ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.3 | 6.4 | REACTOME_HS_GAG_DEGRADATION | Genes involved in HS-GAG degradation |
| 0.3 | 5.6 | REACTOME_AMINE_COMPOUND_SLC_TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.3 | 3.8 | REACTOME_REPAIR_SYNTHESIS_FOR_GAP_FILLING_BY_DNA_POL_IN_TC_NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.3 | 7.8 | REACTOME_G_BETA_GAMMA_SIGNALLING_THROUGH_PLC_BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.3 | 2.3 | REACTOME_BILE_SALT_AND_ORGANIC_ANION_SLC_TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.2 | 6.9 | REACTOME_PRE_NOTCH_TRANSCRIPTION_AND_TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.2 | 2.4 | REACTOME_FORMATION_OF_FIBRIN_CLOT_CLOTTING_CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.1 | 2.0 | REACTOME_INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.1 | 2.0 | REACTOME_ERKS_ARE_INACTIVATED | Genes involved in ERKs are inactivated |
| 0.1 | 3.6 | REACTOME_RNA_POL_I_TRANSCRIPTION_INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.1 | 1.3 | REACTOME_AMINE_LIGAND_BINDING_RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 1.4 | REACTOME_G1_S_SPECIFIC_TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.1 | 7.6 | REACTOME_PEPTIDE_LIGAND_BINDING_RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 1.2 | REACTOME_ROLE_OF_DCC_IN_REGULATING_APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 2.5 | REACTOME_KINESINS | Genes involved in Kinesins |
| 0.1 | 4.1 | REACTOME_INTERFERON_GAMMA_SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 2.0 | REACTOME_CELL_EXTRACELLULAR_MATRIX_INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 1.0 | REACTOME_EXTENSION_OF_TELOMERES | Genes involved in Extension of Telomeres |
| 0.1 | 0.8 | REACTOME_FACILITATIVE_NA_INDEPENDENT_GLUCOSE_TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 4.6 | REACTOME_NUCLEAR_RECEPTOR_TRANSCRIPTION_PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 1.8 | REACTOME_DEADENYLATION_OF_MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 1.0 | REACTOME_MITOCHONDRIAL_FATTY_ACID_BETA_OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 0.4 | REACTOME_INHIBITION_OF_INSULIN_SECRETION_BY_ADRENALINE_NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.1 | 3.0 | REACTOME_NETRIN1_SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 1.6 | REACTOME_REGULATION_OF_WATER_BALANCE_BY_RENAL_AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.1 | 2.7 | REACTOME_CLASS_B_2_SECRETIN_FAMILY_RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 1.0 | REACTOME_RIP_MEDIATED_NFKB_ACTIVATION_VIA_DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 7.3 | REACTOME_TRANSLATION | Genes involved in Translation |
| 0.0 | 0.8 | REACTOME_YAP1_AND_WWTR1_TAZ_STIMULATED_GENE_EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 1.5 | REACTOME_RECYCLING_PATHWAY_OF_L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 1.7 | REACTOME_MITOCHONDRIAL_PROTEIN_IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 1.7 | REACTOME_ACTIVATION_OF_CHAPERONE_GENES_BY_XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.9 | REACTOME_GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 2.0 | REACTOME_NRAGE_SIGNALS_DEATH_THROUGH_JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 1.0 | REACTOME_SPHINGOLIPID_DE_NOVO_BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 1.3 | REACTOME_COLLAGEN_FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.4 | REACTOME_GLYCOGEN_BREAKDOWN_GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.3 | REACTOME_ACTIVATED_NOTCH1_TRANSMITS_SIGNAL_TO_THE_NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.4 | REACTOME_REGULATION_OF_GENE_EXPRESSION_IN_BETA_CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.7 | REACTOME_CDK_MEDIATED_PHOSPHORYLATION_AND_REMOVAL_OF_CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |
| 0.0 | 0.2 | REACTOME_BMAL1_CLOCK_NPAS2_ACTIVATES_CIRCADIAN_EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.2 | REACTOME_HOMOLOGOUS_RECOMBINATION_REPAIR_OF_REPLICATION_INDEPENDENT_DOUBLE_STRAND_BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |