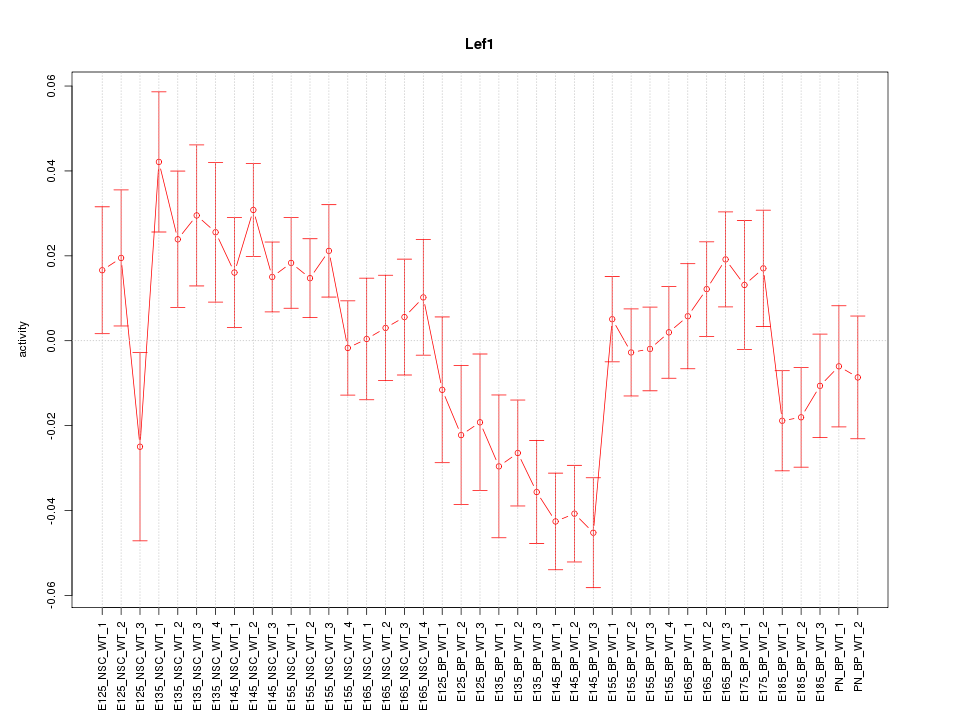
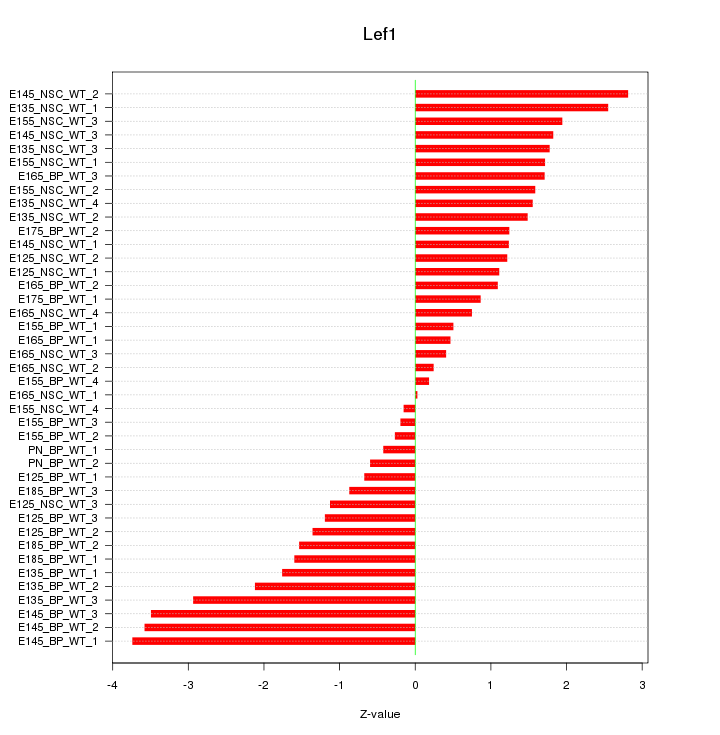
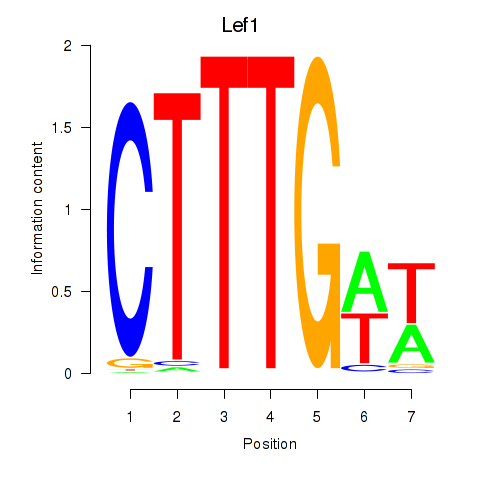
Motif ID: Lef1
Z-value: 1.662
Transcription factors associated with Lef1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Lef1 | ENSMUSG00000027985.8 | Lef1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Lef1 | mm10_v2_chr3_+_131110350_131110471 | 0.22 | 1.6e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 16.9 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 4.2 | 29.2 | GO:0072257 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 3.9 | 11.6 | GO:0061181 | regulation of chondrocyte development(GO:0061181) |
| 3.7 | 11.1 | GO:0061075 | cerebral cortex GABAergic interneuron fate commitment(GO:0021893) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) positive regulation of amacrine cell differentiation(GO:1902871) |
| 2.4 | 7.3 | GO:0046022 | regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 2.3 | 18.3 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 1.7 | 7.0 | GO:2000383 | regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 1.7 | 5.1 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 1.6 | 4.7 | GO:2000314 | negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 1.4 | 5.6 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 1.4 | 10.8 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 1.4 | 4.1 | GO:0048687 | positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) |
| 1.3 | 6.5 | GO:0060242 | contact inhibition(GO:0060242) |
| 1.2 | 3.6 | GO:1902256 | endocardial cushion fusion(GO:0003274) apoptotic process involved in outflow tract morphogenesis(GO:0003275) atrial septum primum morphogenesis(GO:0003289) positive regulation of catagen(GO:0051795) regulation of apoptotic process involved in outflow tract morphogenesis(GO:1902256) |
| 1.1 | 3.4 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 1.1 | 2.1 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 1.0 | 3.9 | GO:0046552 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.9 | 2.8 | GO:0045004 | DNA replication proofreading(GO:0045004) |
| 0.9 | 10.5 | GO:0048853 | forebrain morphogenesis(GO:0048853) |
| 0.8 | 11.9 | GO:2000739 | regulation of mesenchymal stem cell differentiation(GO:2000739) |
| 0.8 | 2.5 | GO:1904020 | regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.8 | 2.5 | GO:0021972 | corticospinal neuron axon guidance through spinal cord(GO:0021972) positive regulation of negative chemotaxis(GO:0050924) induction of negative chemotaxis(GO:0050929) negative regulation of mononuclear cell migration(GO:0071676) negative regulation of retinal ganglion cell axon guidance(GO:0090260) |
| 0.8 | 1.6 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.8 | 3.9 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.8 | 3.1 | GO:0007056 | spindle assembly involved in female meiosis(GO:0007056) |
| 0.8 | 12.2 | GO:0001573 | ganglioside metabolic process(GO:0001573) |
| 0.8 | 3.0 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.8 | 12.1 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.7 | 11.2 | GO:0097154 | GABAergic neuron differentiation(GO:0097154) |
| 0.7 | 2.2 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.7 | 2.7 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.7 | 8.6 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.7 | 2.0 | GO:0061153 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.6 | 14.4 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.5 | 5.4 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.5 | 2.6 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.5 | 0.5 | GO:0071336 | regulation of hair follicle cell proliferation(GO:0071336) positive regulation of hair follicle cell proliferation(GO:0071338) |
| 0.5 | 1.5 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.4 | 2.2 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.4 | 1.6 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.4 | 1.5 | GO:0071699 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.4 | 1.1 | GO:1990927 | negative regulation of membrane invagination(GO:1905154) calcium ion regulated lysosome exocytosis(GO:1990927) |
| 0.4 | 1.1 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.4 | 1.1 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 0.4 | 2.5 | GO:0000320 | re-entry into mitotic cell cycle(GO:0000320) |
| 0.4 | 1.4 | GO:2000182 | regulation of progesterone biosynthetic process(GO:2000182) |
| 0.3 | 3.7 | GO:0002053 | positive regulation of mesenchymal cell proliferation(GO:0002053) |
| 0.3 | 2.3 | GO:0060346 | bone trabecula formation(GO:0060346) |
| 0.3 | 3.3 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.3 | 0.9 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.3 | 1.5 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.3 | 0.6 | GO:2001160 | regulation of histone H3-K79 methylation(GO:2001160) |
| 0.3 | 2.7 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.3 | 1.2 | GO:0002572 | pro-T cell differentiation(GO:0002572) |
| 0.3 | 2.1 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.3 | 1.4 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.3 | 6.7 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.3 | 0.8 | GO:0019405 | alditol catabolic process(GO:0019405) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.3 | 2.2 | GO:0033184 | positive regulation of histone ubiquitination(GO:0033184) |
| 0.3 | 0.8 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.3 | 2.2 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.3 | 1.9 | GO:0070494 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.3 | 2.4 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.3 | 4.5 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.2 | 0.7 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 0.2 | 2.3 | GO:0061470 | interleukin-21 production(GO:0032625) T follicular helper cell differentiation(GO:0061470) interleukin-21 secretion(GO:0072619) |
| 0.2 | 3.4 | GO:0015809 | arginine transport(GO:0015809) |
| 0.2 | 1.5 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.2 | 0.8 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 0.2 | 1.4 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.2 | 3.9 | GO:0030903 | notochord development(GO:0030903) |
| 0.2 | 1.9 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.2 | 0.5 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.2 | 0.6 | GO:0050904 | diapedesis(GO:0050904) |
| 0.2 | 1.3 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 | 1.3 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.1 | 0.4 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.1 | 0.4 | GO:0071579 | regulation of zinc ion transport(GO:0071579) |
| 0.1 | 7.6 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.1 | 2.5 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.1 | 0.6 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.1 | 0.6 | GO:0003097 | renal water transport(GO:0003097) renal water absorption(GO:0070295) |
| 0.1 | 0.1 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.1 | 3.2 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.1 | 1.3 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.1 | 1.0 | GO:2000794 | regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000794) |
| 0.1 | 0.6 | GO:2000325 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.1 | 4.1 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.1 | 13.2 | GO:0030326 | embryonic limb morphogenesis(GO:0030326) embryonic appendage morphogenesis(GO:0035113) |
| 0.1 | 1.4 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.1 | 0.7 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.1 | 0.5 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.1 | 0.5 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 0.6 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) establishment of Sertoli cell barrier(GO:0097368) |
| 0.1 | 0.8 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.1 | 0.8 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.1 | 0.3 | GO:1904207 | chemokine (C-C motif) ligand 2 secretion(GO:0035926) regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.1 | 0.3 | GO:1904245 | regulation of polynucleotide adenylyltransferase activity(GO:1904245) |
| 0.1 | 0.7 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.1 | 3.7 | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.1 | 1.7 | GO:0001967 | suckling behavior(GO:0001967) |
| 0.1 | 2.4 | GO:0095500 | acetylcholine receptor signaling pathway(GO:0095500) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.1 | 1.7 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.1 | 1.1 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.1 | 1.2 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.1 | 8.7 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.1 | 1.1 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.1 | 1.0 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.1 | 0.3 | GO:0033580 | protein glycosylation at cell surface(GO:0033575) protein galactosylation at cell surface(GO:0033580) protein galactosylation(GO:0042125) |
| 0.1 | 0.4 | GO:0097473 | cellular response to light intensity(GO:0071484) cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.1 | 0.9 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.1 | 0.9 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 0.3 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.1 | 0.7 | GO:0010763 | positive regulation of fibroblast migration(GO:0010763) |
| 0.1 | 0.9 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.1 | 3.0 | GO:0060976 | coronary vasculature development(GO:0060976) |
| 0.1 | 0.4 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.1 | 0.9 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.0 | 0.2 | GO:0035547 | interferon-beta secretion(GO:0035546) regulation of interferon-beta secretion(GO:0035547) positive regulation of interferon-beta secretion(GO:0035549) CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.3 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 2.7 | GO:0001885 | endothelial cell development(GO:0001885) |
| 0.0 | 1.3 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.2 | GO:0071442 | N-terminal peptidyl-lysine acetylation(GO:0018076) regulation of histone H3-K14 acetylation(GO:0071440) positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.0 | 1.6 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 | 0.2 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.0 | 4.8 | GO:0006261 | DNA-dependent DNA replication(GO:0006261) |
| 0.0 | 0.3 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.1 | GO:0003263 | cardioblast proliferation(GO:0003263) regulation of cardioblast proliferation(GO:0003264) vitamin D receptor signaling pathway(GO:0070561) positive regulation of cell proliferation involved in heart morphogenesis(GO:2000138) |
| 0.0 | 0.3 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 1.0 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.8 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.0 | 0.2 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 0.1 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 1.0 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 1.3 | GO:0045638 | negative regulation of myeloid cell differentiation(GO:0045638) |
| 0.0 | 2.3 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.2 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.4 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 0.2 | GO:0060712 | spongiotrophoblast layer development(GO:0060712) |
| 0.0 | 0.2 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.2 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 0.9 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.6 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.0 | 1.1 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.2 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 2.0 | GO:0090132 | epithelial cell migration(GO:0010631) tissue migration(GO:0090130) epithelium migration(GO:0090132) |
| 0.0 | 0.1 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 0.5 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.3 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.1 | GO:0061436 | establishment of skin barrier(GO:0061436) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 18.3 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 1.2 | 4.8 | GO:0008623 | CHRAC(GO:0008623) |
| 1.2 | 3.6 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.8 | 3.2 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.6 | 3.1 | GO:0005587 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.6 | 1.8 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.6 | 1.8 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.6 | 1.2 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.6 | 13.1 | GO:0008305 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.6 | 2.8 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.5 | 8.5 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.5 | 3.9 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.4 | 2.8 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.4 | 8.6 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.3 | 1.7 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.2 | 0.9 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.2 | 2.8 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.2 | 3.3 | GO:0042555 | MCM complex(GO:0042555) |
| 0.2 | 11.5 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.2 | 1.1 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.2 | 72.4 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.2 | 0.6 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.2 | 5.8 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 2.4 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.1 | 1.1 | GO:0032009 | early phagosome(GO:0032009) |
| 0.1 | 1.5 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.1 | 1.7 | GO:0031105 | septin complex(GO:0031105) |
| 0.1 | 1.4 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.1 | 17.2 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.1 | 1.6 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 0.9 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 0.3 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.1 | 0.7 | GO:0042581 | specific granule(GO:0042581) |
| 0.1 | 0.2 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.1 | 0.9 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.5 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 4.7 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.2 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.0 | 4.1 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 1.7 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 1.4 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 1.5 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.4 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 2.7 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.7 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.1 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.0 | 0.2 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 1.2 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.4 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 2.0 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 1.6 | GO:0030118 | clathrin coat(GO:0030118) |
| 0.0 | 1.2 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.3 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.2 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.6 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.1 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.2 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 1.0 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.3 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 2.2 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.1 | GO:0005677 | chromatin silencing complex(GO:0005677) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 16.9 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 1.9 | 13.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 1.5 | 12.3 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.9 | 2.8 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.9 | 19.5 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.9 | 3.6 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.8 | 4.1 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.8 | 2.4 | GO:0030549 | acetylcholine receptor activator activity(GO:0030549) |
| 0.8 | 5.6 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) lysophosphatidic acid receptor activity(GO:0070915) |
| 0.8 | 2.3 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.6 | 5.1 | GO:0005283 | sodium:amino acid symporter activity(GO:0005283) |
| 0.6 | 11.1 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.6 | 2.8 | GO:0005113 | patched binding(GO:0005113) |
| 0.5 | 3.0 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.5 | 1.9 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.5 | 1.9 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 0.5 | 3.7 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.4 | 2.2 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.4 | 3.4 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.4 | 3.4 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.4 | 5.0 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.4 | 5.7 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.4 | 2.2 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.3 | 5.7 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.3 | 1.3 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.3 | 8.7 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.3 | 1.3 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.3 | 4.1 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.3 | 4.0 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.3 | 7.0 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.3 | 1.9 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.3 | 1.3 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.3 | 2.3 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.3 | 6.0 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.2 | 7.4 | GO:0042805 | actinin binding(GO:0042805) |
| 0.2 | 1.1 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.2 | 0.9 | GO:0003844 | 1,4-alpha-glucan branching enzyme activity(GO:0003844) |
| 0.2 | 3.6 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.2 | 0.8 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.2 | 4.8 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.2 | 0.9 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.2 | 8.6 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.2 | 0.7 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.2 | 8.6 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.2 | 1.4 | GO:0043559 | insulin binding(GO:0043559) |
| 0.2 | 3.4 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.2 | 4.0 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.2 | 32.5 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.2 | 1.1 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.2 | 22.6 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 0.4 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.1 | 2.1 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 0.5 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.1 | 3.9 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 0.9 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 2.6 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 7.1 | GO:0000987 | core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.1 | 0.8 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 1.2 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.1 | 0.6 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 0.5 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.1 | 3.5 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.1 | 3.7 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 0.8 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.1 | 2.0 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 40.4 | GO:0000981 | RNA polymerase II transcription factor activity, sequence-specific DNA binding(GO:0000981) |
| 0.1 | 1.4 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 0.3 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.1 | 0.7 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.1 | 1.2 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 8.8 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.3 | GO:0047276 | N-acetyllactosaminide 3-alpha-galactosyltransferase activity(GO:0047276) |
| 0.0 | 0.9 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.1 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.9 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 14.5 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 0.2 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.8 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.3 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.3 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 0.5 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 1.0 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 1.7 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.2 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.6 | GO:0019903 | protein phosphatase binding(GO:0019903) |
| 0.0 | 1.1 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.1 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.8 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.2 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 1.8 | GO:0003700 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 0.2 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 1.1 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.4 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 13.1 | PID_INTEGRIN5_PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.5 | 55.0 | PID_BETA_CATENIN_NUC_PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.4 | 3.6 | SA_MMP_CYTOKINE_CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.2 | 7.0 | PID_SYNDECAN_4_PATHWAY | Syndecan-4-mediated signaling events |
| 0.2 | 4.5 | ST_G_ALPHA_S_PATHWAY | G alpha s Pathway |
| 0.1 | 2.0 | PID_SYNDECAN_1_PATHWAY | Syndecan-1-mediated signaling events |
| 0.1 | 13.0 | PID_SMAD2_3NUCLEAR_PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.1 | 6.5 | PID_BMP_PATHWAY | BMP receptor signaling |
| 0.1 | 2.5 | PID_GLYPICAN_1PATHWAY | Glypican 1 network |
| 0.1 | 3.3 | PID_ERBB2_ERBB3_PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 3.1 | PID_INTEGRIN3_PATHWAY | Beta3 integrin cell surface interactions |
| 0.1 | 2.7 | PID_HEDGEHOG_2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 5.6 | PID_PLK1_PATHWAY | PLK1 signaling events |
| 0.1 | 4.1 | PID_HES_HEY_PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 1.8 | PID_IL12_STAT4_PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 3.0 | NABA_COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 2.9 | PID_ATF2_PATHWAY | ATF-2 transcription factor network |
| 0.1 | 5.3 | PID_LYSOPHOSPHOLIPID_PATHWAY | LPA receptor mediated events |
| 0.1 | 2.3 | PID_SHP2_PATHWAY | SHP2 signaling |
| 0.1 | 2.7 | PID_LKB1_PATHWAY | LKB1 signaling events |
| 0.0 | 1.7 | PID_ATR_PATHWAY | ATR signaling pathway |
| 0.0 | 0.4 | SA_TRKA_RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 2.2 | PID_RAC1_PATHWAY | RAC1 signaling pathway |
| 0.0 | 2.1 | PID_E2F_PATHWAY | E2F transcription factor network |
| 0.0 | 0.7 | PID_P38_ALPHA_BETA_PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 1.2 | PID_FAK_PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.8 | PID_WNT_NONCANONICAL_PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 3.2 | NABA_ECM_GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.1 | PID_IL5_PATHWAY | IL5-mediated signaling events |
| 0.0 | 1.1 | PID_P53_REGULATION_PATHWAY | p53 pathway |
| 0.0 | 0.7 | PID_ARF6_TRAFFICKING_PATHWAY | Arf6 trafficking events |
| 0.0 | 2.1 | NABA_ECM_REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.5 | PID_AR_TF_PATHWAY | Regulation of Androgen receptor activity |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 32.9 | REACTOME_YAP1_AND_WWTR1_TAZ_STIMULATED_GENE_EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.5 | 3.2 | REACTOME_TRANSLOCATION_OF_ZAP_70_TO_IMMUNOLOGICAL_SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.5 | 5.1 | REACTOME_NA_CL_DEPENDENT_NEUROTRANSMITTER_TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.5 | 2.3 | REACTOME_SIGNALING_BY_FGFR | Genes involved in Signaling by FGFR |
| 0.4 | 19.0 | REACTOME_AMINO_ACID_AND_OLIGOPEPTIDE_SLC_TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.3 | 4.8 | REACTOME_CONVERSION_FROM_APC_C_CDC20_TO_APC_C_CDH1_IN_LATE_ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.2 | 3.3 | REACTOME_UNWINDING_OF_DNA | Genes involved in Unwinding of DNA |
| 0.2 | 2.3 | REACTOME_SIGNALING_BY_PDGF | Genes involved in Signaling by PDGF |
| 0.2 | 2.8 | REACTOME_REPAIR_SYNTHESIS_FOR_GAP_FILLING_BY_DNA_POL_IN_TC_NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.2 | 15.0 | REACTOME_INTEGRIN_CELL_SURFACE_INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.2 | 2.5 | REACTOME_ACTIVATION_OF_RAC | Genes involved in Activation of Rac |
| 0.2 | 1.4 | REACTOME_CIRCADIAN_REPRESSION_OF_EXPRESSION_BY_REV_ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.2 | 1.1 | REACTOME_TANDEM_PORE_DOMAIN_POTASSIUM_CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.1 | 3.9 | REACTOME_PRE_NOTCH_TRANSCRIPTION_AND_TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.1 | 5.2 | REACTOME_DOWNREGULATION_OF_SMAD2_3_SMAD4_TRANSCRIPTIONAL_ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.1 | 1.9 | REACTOME_G1_S_SPECIFIC_TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.1 | 3.0 | REACTOME_SIGNALING_BY_BMP | Genes involved in Signaling by BMP |
| 0.1 | 2.6 | REACTOME_DOWNREGULATION_OF_TGF_BETA_RECEPTOR_SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.1 | 3.0 | REACTOME_SULFUR_AMINO_ACID_METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 8.3 | REACTOME_CLASS_B_2_SECRETIN_FAMILY_RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 0.7 | REACTOME_REGULATION_OF_INSULIN_SECRETION_BY_ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 0.6 | REACTOME_PASSIVE_TRANSPORT_BY_AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 3.1 | REACTOME_RNA_POL_III_TRANSCRIPTION | Genes involved in RNA Polymerase III Transcription |
| 0.1 | 4.2 | REACTOME_NUCLEAR_RECEPTOR_TRANSCRIPTION_PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 0.9 | REACTOME_VITAMIN_B5_PANTOTHENATE_METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 1.6 | REACTOME_KINESINS | Genes involved in Kinesins |
| 0.1 | 6.1 | REACTOME_MEIOSIS | Genes involved in Meiosis |
| 0.1 | 5.5 | REACTOME_G_ALPHA_Q_SIGNALLING_EVENTS | Genes involved in G alpha (q) signalling events |
| 0.1 | 0.4 | REACTOME_G_BETA_GAMMA_SIGNALLING_THROUGH_PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 1.3 | REACTOME_SIGNALING_BY_FGFR1_FUSION_MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 1.9 | REACTOME_SEMA4D_IN_SEMAPHORIN_SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 0.6 | REACTOME_G0_AND_EARLY_G1 | Genes involved in G0 and Early G1 |
| 0.0 | 1.9 | REACTOME_COLLAGEN_FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.5 | REACTOME_ACYL_CHAIN_REMODELLING_OF_PC | Genes involved in Acyl chain remodelling of PC |
| 0.0 | 1.1 | REACTOME_ADHERENS_JUNCTIONS_INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.1 | REACTOME_IL_7_SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 1.2 | REACTOME_NETRIN1_SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.5 | REACTOME_PROCESSING_OF_INTRONLESS_PRE_MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 2.2 | REACTOME_FACTORS_INVOLVED_IN_MEGAKARYOCYTE_DEVELOPMENT_AND_PLATELET_PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.4 | REACTOME_ZINC_TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.8 | REACTOME_GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.9 | REACTOME_GLUCOSE_METABOLISM | Genes involved in Glucose metabolism |
| 0.0 | 1.6 | REACTOME_RESPONSE_TO_ELEVATED_PLATELET_CYTOSOLIC_CA2_ | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.4 | REACTOME_CHONDROITIN_SULFATE_BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 1.1 | REACTOME_APC_C_CDH1_MEDIATED_DEGRADATION_OF_CDC20_AND_OTHER_APC_C_CDH1_TARGETED_PROTEINS_IN_LATE_MITOSIS_EARLY_G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.0 | 0.8 | REACTOME_APOPTOTIC_CLEAVAGE_OF_CELLULAR_PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.2 | REACTOME_SYNTHESIS_SECRETION_AND_INACTIVATION_OF_GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 0.9 | REACTOME_GOLGI_ASSOCIATED_VESICLE_BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |