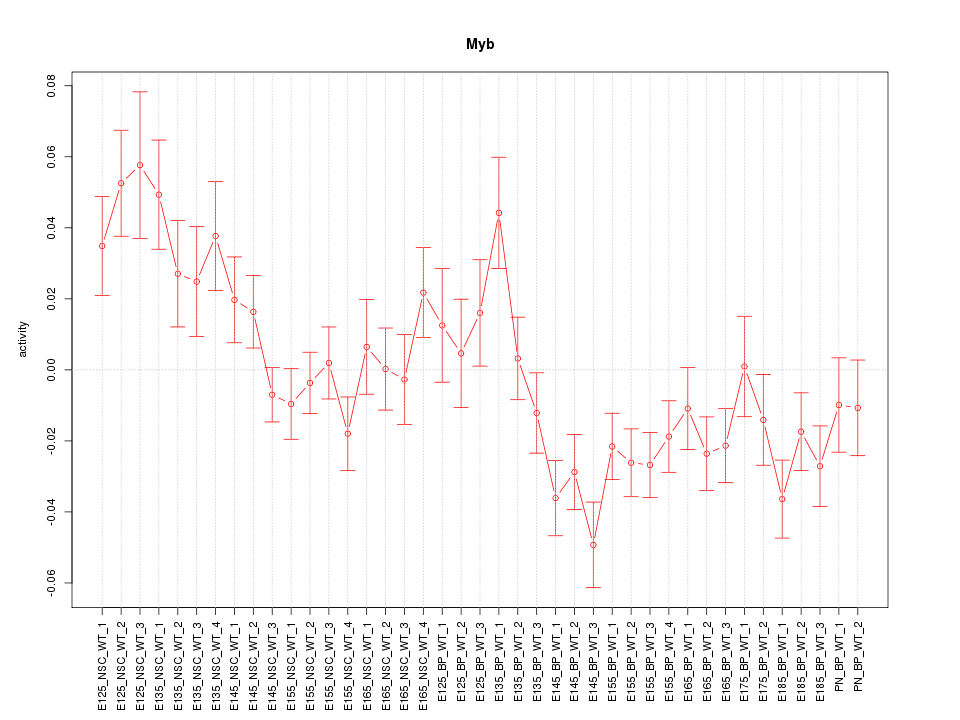
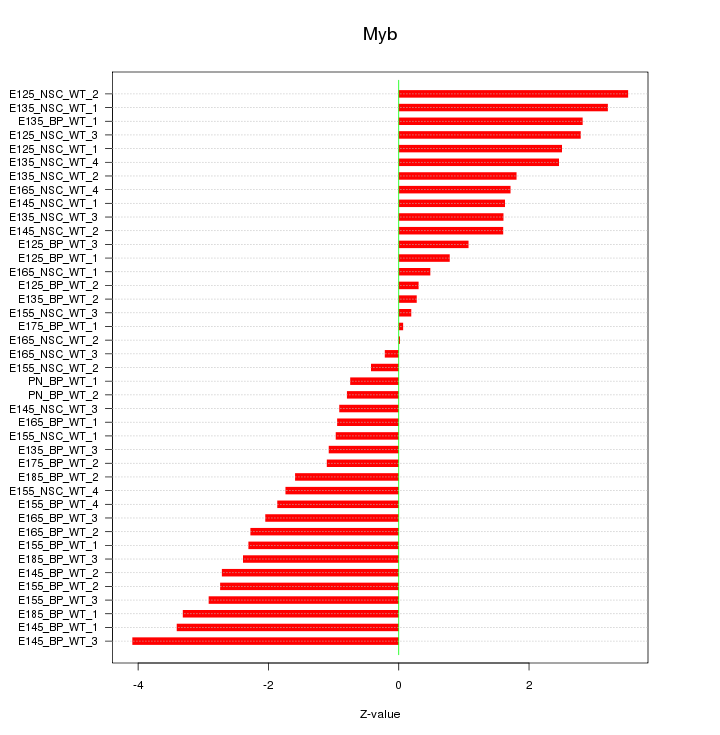
Motif ID: Myb
Z-value: 2.010
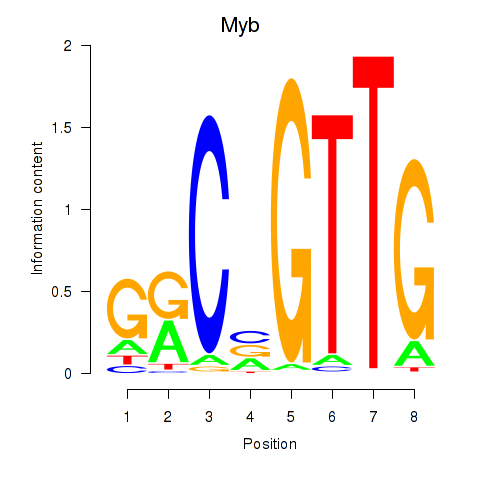
Transcription factors associated with Myb:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Myb | ENSMUSG00000019982.8 | Myb |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Myb | mm10_v2_chr10_-_21160925_21160984 | 0.84 | 6.6e-12 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.2 | 37.1 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 3.6 | 10.9 | GO:0072382 | minus-end-directed vesicle transport along microtubule(GO:0072382) |
| 2.9 | 8.6 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 2.1 | 10.3 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 2.0 | 36.1 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 2.0 | 10.0 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 1.9 | 5.8 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 1.6 | 11.3 | GO:0000279 | M phase(GO:0000279) |
| 1.4 | 8.4 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 1.3 | 17.2 | GO:0099628 | receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 1.3 | 7.9 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 1.3 | 7.5 | GO:1901911 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 1.2 | 8.4 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 1.0 | 2.0 | GO:0072717 | cellular response to actinomycin D(GO:0072717) |
| 1.0 | 8.0 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 1.0 | 11.8 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 1.0 | 16.3 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.9 | 4.7 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 0.9 | 3.7 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.9 | 2.8 | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.9 | 4.6 | GO:0019659 | fermentation(GO:0006113) lactate biosynthetic process from pyruvate(GO:0019244) glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.8 | 1.7 | GO:0031660 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) |
| 0.8 | 5.9 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.8 | 5.8 | GO:0000320 | re-entry into mitotic cell cycle(GO:0000320) |
| 0.8 | 13.2 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.7 | 3.6 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.7 | 7.0 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.7 | 4.1 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.6 | 5.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.6 | 3.8 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.6 | 1.9 | GO:2000564 | CD8-positive, alpha-beta T cell proliferation(GO:0035740) negative regulation of regulatory T cell differentiation(GO:0045590) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) |
| 0.6 | 2.3 | GO:1990022 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.6 | 8.9 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.5 | 6.9 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.5 | 1.5 | GO:1901536 | negative regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045869) regulation of DNA demethylation(GO:1901535) negative regulation of DNA demethylation(GO:1901536) |
| 0.5 | 2.5 | GO:0033762 | response to glucagon(GO:0033762) |
| 0.5 | 1.5 | GO:1901420 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) negative regulation of response to alcohol(GO:1901420) positive regulation of macrophage apoptotic process(GO:2000111) |
| 0.5 | 2.4 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.5 | 1.9 | GO:0019355 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.5 | 3.8 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.5 | 1.4 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.4 | 1.3 | GO:0009826 | unidimensional cell growth(GO:0009826) susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.4 | 2.2 | GO:0032472 | Golgi calcium ion transport(GO:0032472) |
| 0.4 | 7.8 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.4 | 1.3 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.4 | 1.2 | GO:0014028 | notochord formation(GO:0014028) |
| 0.4 | 2.0 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.4 | 1.2 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.4 | 2.8 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.4 | 8.6 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.4 | 1.1 | GO:0090202 | transcriptional activation by promoter-enhancer looping(GO:0071733) regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.4 | 1.1 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.3 | 2.1 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) choline metabolic process(GO:0019695) |
| 0.3 | 1.0 | GO:0009174 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.3 | 1.0 | GO:0098968 | neurotransmitter receptor transport postsynaptic membrane to endosome(GO:0098968) |
| 0.3 | 9.7 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.3 | 2.9 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.3 | 1.9 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.3 | 1.2 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.3 | 0.9 | GO:0032241 | positive regulation of nucleobase-containing compound transport(GO:0032241) |
| 0.3 | 1.0 | GO:0033600 | negative regulation of mammary gland epithelial cell proliferation(GO:0033600) |
| 0.2 | 2.7 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.2 | 1.0 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.2 | 8.0 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.2 | 1.6 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.2 | 0.9 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.2 | 2.3 | GO:1990173 | protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) protein localization to nucleoplasm(GO:1990173) |
| 0.2 | 1.1 | GO:2001170 | negative regulation of ATP biosynthetic process(GO:2001170) |
| 0.2 | 4.0 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.2 | 1.8 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.2 | 1.4 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.2 | 1.2 | GO:0009169 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.2 | 3.3 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.2 | 4.1 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.2 | 1.2 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.2 | 5.5 | GO:0034508 | centromere complex assembly(GO:0034508) |
| 0.2 | 1.4 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.2 | 1.7 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.2 | 0.8 | GO:0002667 | lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) negative regulation of protein kinase C signaling(GO:0090038) |
| 0.2 | 1.8 | GO:0035036 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) |
| 0.2 | 1.6 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.2 | 1.0 | GO:1902965 | regulation of protein localization to early endosome(GO:1902965) positive regulation of protein localization to early endosome(GO:1902966) |
| 0.2 | 2.6 | GO:0046599 | regulation of centriole replication(GO:0046599) |
| 0.1 | 6.3 | GO:0048255 | mRNA stabilization(GO:0048255) |
| 0.1 | 2.1 | GO:0007099 | centriole replication(GO:0007099) |
| 0.1 | 1.2 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.1 | 7.2 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.1 | 2.3 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.1 | 2.5 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.1 | 2.8 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.1 | 0.9 | GO:0002643 | regulation of tolerance induction(GO:0002643) |
| 0.1 | 1.3 | GO:0046051 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.1 | 1.0 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.1 | 1.9 | GO:2001273 | regulation of glucose import in response to insulin stimulus(GO:2001273) |
| 0.1 | 0.6 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.1 | 0.8 | GO:0043634 | polyadenylation-dependent ncRNA catabolic process(GO:0043634) |
| 0.1 | 1.1 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.1 | 16.1 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.1 | 0.5 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.1 | 0.3 | GO:0002014 | vasoconstriction of artery involved in ischemic response to lowering of systemic arterial blood pressure(GO:0002014) |
| 0.1 | 0.9 | GO:0002155 | regulation of thyroid hormone mediated signaling pathway(GO:0002155) |
| 0.1 | 3.5 | GO:1902750 | negative regulation of cell cycle G2/M phase transition(GO:1902750) |
| 0.1 | 0.5 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.1 | 1.1 | GO:0044380 | protein localization to cytoskeleton(GO:0044380) |
| 0.1 | 0.3 | GO:0090238 | positive regulation of arachidonic acid secretion(GO:0090238) |
| 0.1 | 6.7 | GO:0007093 | mitotic cell cycle checkpoint(GO:0007093) |
| 0.1 | 0.7 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.1 | 0.3 | GO:0034162 | toll-like receptor 9 signaling pathway(GO:0034162) |
| 0.1 | 0.6 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.1 | 1.2 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.1 | 4.4 | GO:0007127 | meiosis I(GO:0007127) |
| 0.1 | 1.0 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.1 | 0.5 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.1 | 1.1 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.1 | 0.7 | GO:0022409 | positive regulation of cell-cell adhesion(GO:0022409) |
| 0.0 | 1.2 | GO:2000060 | positive regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000060) |
| 0.0 | 0.9 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 0.3 | GO:0018377 | N-terminal protein myristoylation(GO:0006499) protein myristoylation(GO:0018377) |
| 0.0 | 0.9 | GO:0000469 | cleavage involved in rRNA processing(GO:0000469) |
| 0.0 | 0.3 | GO:0035701 | hematopoietic stem cell migration(GO:0035701) |
| 0.0 | 1.0 | GO:2000780 | negative regulation of DNA repair(GO:0045738) negative regulation of double-strand break repair(GO:2000780) |
| 0.0 | 0.4 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.0 | 0.4 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 1.4 | GO:0034766 | negative regulation of ion transmembrane transport(GO:0034766) |
| 0.0 | 0.2 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 2.2 | GO:0000070 | mitotic sister chromatid segregation(GO:0000070) |
| 0.0 | 1.6 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 1.2 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 3.1 | GO:0008584 | male gonad development(GO:0008584) development of primary male sexual characteristics(GO:0046546) |
| 0.0 | 3.3 | GO:0048663 | neuron fate commitment(GO:0048663) |
| 0.0 | 0.3 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.0 | 0.1 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 | 3.0 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 0.7 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 2.0 | GO:0000271 | polysaccharide biosynthetic process(GO:0000271) |
| 0.0 | 4.1 | GO:0000082 | G1/S transition of mitotic cell cycle(GO:0000082) |
| 0.0 | 0.8 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.1 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 | 1.5 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.7 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 1.5 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.6 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 1.1 | GO:0030500 | regulation of bone mineralization(GO:0030500) |
| 0.0 | 0.7 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.8 | GO:0006970 | response to osmotic stress(GO:0006970) |
| 0.0 | 0.7 | GO:0046324 | regulation of glucose import(GO:0046324) |
| 0.0 | 0.4 | GO:0002437 | inflammatory response to antigenic stimulus(GO:0002437) |
| 0.0 | 0.1 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.0 | 1.5 | GO:0030218 | erythrocyte differentiation(GO:0030218) |
| 0.0 | 0.3 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 1.7 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 0.3 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.0 | 1.7 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.0 | 2.3 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 1.2 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.2 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.0 | 0.3 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.3 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 1.1 | GO:0006364 | rRNA processing(GO:0006364) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.9 | 17.8 | GO:0099573 | glutamatergic postsynaptic density(GO:0099573) |
| 3.4 | 37.5 | GO:0005642 | annulate lamellae(GO:0005642) |
| 2.2 | 6.7 | GO:1990423 | RZZ complex(GO:1990423) |
| 2.1 | 8.6 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 1.9 | 5.8 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.9 | 6.3 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.9 | 32.0 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.8 | 2.3 | GO:1990047 | spindle matrix(GO:1990047) |
| 0.7 | 5.1 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.7 | 5.0 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.7 | 4.8 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.7 | 1.4 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.6 | 4.5 | GO:0060293 | P granule(GO:0043186) pole plasm(GO:0045495) germ plasm(GO:0060293) |
| 0.6 | 1.9 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.6 | 10.9 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.6 | 2.4 | GO:1990590 | ATF1-ATF4 transcription factor complex(GO:1990590) |
| 0.6 | 4.1 | GO:0001740 | Barr body(GO:0001740) |
| 0.6 | 3.3 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.5 | 2.0 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.5 | 4.5 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.5 | 1.5 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.5 | 1.4 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.4 | 10.9 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.4 | 1.2 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.4 | 0.4 | GO:0099524 | region of cytosol(GO:0099522) postsynaptic cytosol(GO:0099524) |
| 0.4 | 1.9 | GO:0097226 | sperm mitochondrial sheath(GO:0097226) |
| 0.4 | 8.4 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.3 | 6.7 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.3 | 4.6 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.3 | 1.5 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.3 | 1.8 | GO:0098536 | deuterosome(GO:0098536) |
| 0.3 | 0.9 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.3 | 4.1 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.3 | 2.7 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.2 | 2.4 | GO:0000243 | commitment complex(GO:0000243) |
| 0.2 | 14.1 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.2 | 12.2 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.2 | 5.7 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.2 | 3.0 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.2 | 1.0 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.2 | 2.3 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.2 | 5.2 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.2 | 10.8 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.2 | 1.8 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.2 | 15.9 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.2 | 1.1 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.1 | 0.9 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.1 | 0.8 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 11.3 | GO:0000922 | spindle pole(GO:0000922) |
| 0.1 | 8.5 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.1 | 8.5 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 1.8 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.1 | 0.8 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.1 | 10.4 | GO:0000776 | kinetochore(GO:0000776) |
| 0.1 | 1.0 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 9.3 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.1 | 1.9 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.1 | 8.2 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 7.2 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.1 | 2.2 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 4.7 | GO:0005844 | polysome(GO:0005844) |
| 0.1 | 0.5 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.1 | 1.1 | GO:0005605 | basal lamina(GO:0005605) |
| 0.1 | 5.9 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 17.1 | GO:0005813 | centrosome(GO:0005813) |
| 0.0 | 1.0 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 1.2 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 1.5 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 0.3 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 4.0 | GO:0022626 | cytosolic ribosome(GO:0022626) |
| 0.0 | 0.9 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 0.9 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 1.5 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 8.2 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.7 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.6 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.6 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 0.7 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.5 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) small ribosomal subunit(GO:0015935) |
| 0.0 | 13.3 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 2.8 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 1.0 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 2.8 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.8 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.9 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.0 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.0 | 0.5 | GO:0008287 | protein serine/threonine phosphatase complex(GO:0008287) phosphatase complex(GO:1903293) |
| 0.0 | 0.1 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 1.0 | GO:1990204 | oxidoreductase complex(GO:1990204) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 10.3 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 2.3 | 37.1 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 1.9 | 11.3 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 1.7 | 5.1 | GO:0071207 | histone pre-mRNA stem-loop binding(GO:0071207) |
| 1.7 | 6.7 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 1.5 | 17.8 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 1.3 | 5.0 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 1.3 | 7.5 | GO:0000298 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 1.2 | 5.8 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 1.1 | 4.6 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 1.0 | 5.9 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.8 | 3.2 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.6 | 8.4 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.6 | 8.8 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.5 | 3.8 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.5 | 2.1 | GO:0004104 | choline kinase activity(GO:0004103) cholinesterase activity(GO:0004104) choline binding(GO:0033265) |
| 0.5 | 1.5 | GO:0046969 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.5 | 1.9 | GO:0004515 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.4 | 3.6 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.4 | 7.0 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.4 | 1.5 | GO:0035851 | Krueppel-associated box domain binding(GO:0035851) |
| 0.4 | 7.9 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.4 | 2.9 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.4 | 5.0 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.3 | 2.0 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.3 | 10.5 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.3 | 8.6 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.3 | 2.4 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.3 | 2.4 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.2 | 4.7 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.2 | 3.0 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.2 | 1.6 | GO:0032564 | dATP binding(GO:0032564) |
| 0.2 | 5.8 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.2 | 1.0 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.2 | 0.8 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.2 | 6.2 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.2 | 1.0 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 0.2 | 1.1 | GO:0043141 | TFIID-class transcription factor binding(GO:0001094) ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.2 | 8.3 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.2 | 1.9 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.2 | 12.2 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.2 | 0.9 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 10.9 | GO:0004004 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.1 | 1.7 | GO:0016701 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen(GO:0016701) oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.1 | 5.6 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.1 | 1.5 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.1 | 6.0 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.1 | 7.9 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.1 | 1.8 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.1 | 0.8 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.1 | 1.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 3.7 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 1.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 1.1 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.1 | 1.3 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.1 | 1.2 | GO:0031386 | protein tag(GO:0031386) |
| 0.1 | 1.8 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 4.1 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 1.9 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 1.2 | GO:0048038 | quinone binding(GO:0048038) |
| 0.1 | 0.5 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.1 | 0.9 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.1 | 0.1 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.1 | 2.2 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 4.0 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.1 | 0.9 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 1.5 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 0.4 | GO:0070739 | NEDD8 transferase activity(GO:0019788) protein-glutamic acid ligase activity(GO:0070739) |
| 0.1 | 1.9 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.1 | 1.8 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 3.1 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.8 | GO:0051861 | glycolipid binding(GO:0051861) |
| 0.0 | 1.9 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 1.4 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.7 | GO:0015149 | hexose transmembrane transporter activity(GO:0015149) |
| 0.0 | 1.2 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 1.0 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 1.7 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 1.0 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 1.7 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.9 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 6.2 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.5 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 3.0 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 0.8 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.2 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 2.5 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.7 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.7 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 8.4 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 1.6 | GO:0008173 | RNA methyltransferase activity(GO:0008173) |
| 0.0 | 0.9 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.3 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 2.7 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 1.1 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 1.7 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.3 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 1.2 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.2 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.1 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 2.0 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.7 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 0.1 | GO:0030275 | LRR domain binding(GO:0030275) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 17.8 | ST_WNT_CA2_CYCLIC_GMP_PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.6 | 40.2 | PID_PLK1_PATHWAY | PLK1 signaling events |
| 0.5 | 17.0 | PID_NECTIN_PATHWAY | Nectin adhesion pathway |
| 0.4 | 27.8 | PID_ILK_PATHWAY | Integrin-linked kinase signaling |
| 0.3 | 8.6 | PID_SMAD2_3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.2 | 16.6 | PID_TELOMERASE_PATHWAY | Regulation of Telomerase |
| 0.2 | 2.8 | PID_ERB_GENOMIC_PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 4.3 | PID_INSULIN_GLUCOSE_PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 7.9 | PID_SMAD2_3NUCLEAR_PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.1 | 3.0 | PID_ATM_PATHWAY | ATM pathway |
| 0.1 | 3.6 | PID_FOXM1_PATHWAY | FOXM1 transcription factor network |
| 0.1 | 1.2 | PID_TCR_RAS_PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.1 | 2.4 | PID_BETA_CATENIN_DEG_PATHWAY | Degradation of beta catenin |
| 0.1 | 1.5 | PID_HDAC_CLASSIII_PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 1.2 | PID_EPHA2_FWD_PATHWAY | EPHA2 forward signaling |
| 0.1 | 5.0 | PID_AVB3_INTEGRIN_PATHWAY | Integrins in angiogenesis |
| 0.1 | 4.6 | PID_CMYB_PATHWAY | C-MYB transcription factor network |
| 0.1 | 2.4 | ST_P38_MAPK_PATHWAY | p38 MAPK Pathway |
| 0.1 | 4.0 | PID_P53_REGULATION_PATHWAY | p53 pathway |
| 0.1 | 0.5 | PID_TRAIL_PATHWAY | TRAIL signaling pathway |
| 0.1 | 0.9 | PID_P38_MKK3_6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 0.9 | PID_PRL_SIGNALING_EVENTS_PATHWAY | Signaling events mediated by PRL |
| 0.1 | 4.8 | PID_MYC_ACTIV_PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 2.1 | PID_CASPASE_PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 1.1 | PID_AURORA_B_PATHWAY | Aurora B signaling |
| 0.0 | 0.6 | PID_FAK_PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 1.0 | PID_RHOA_PATHWAY | RhoA signaling pathway |
| 0.0 | 1.0 | PID_REG_GR_PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.3 | PID_BCR_5PATHWAY | BCR signaling pathway |
| 0.0 | 0.5 | PID_NETRIN_PATHWAY | Netrin-mediated signaling events |
| 0.0 | 1.3 | WNT_SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 2.1 | NABA_ECM_GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.6 | PID_LKB1_PATHWAY | LKB1 signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 37.1 | REACTOME_PTM_GAMMA_CARBOXYLATION_HYPUSINE_FORMATION_AND_ARYLSULFATASE_ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.8 | 6.7 | REACTOME_APOBEC3G_MEDIATED_RESISTANCE_TO_HIV1_INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.7 | 17.3 | REACTOME_CREB_PHOSPHORYLATION_THROUGH_THE_ACTIVATION_OF_CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.7 | 18.7 | REACTOME_KINESINS | Genes involved in Kinesins |
| 0.6 | 15.0 | REACTOME_APC_CDC20_MEDIATED_DEGRADATION_OF_NEK2A | Genes involved in APC-Cdc20 mediated degradation of Nek2A |
| 0.5 | 18.3 | REACTOME_ADHERENS_JUNCTIONS_INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.4 | 5.1 | REACTOME_SLBP_DEPENDENT_PROCESSING_OF_REPLICATION_DEPENDENT_HISTONE_PRE_MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.4 | 39.4 | REACTOME_MITOTIC_PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.3 | 10.2 | REACTOME_PRE_NOTCH_TRANSCRIPTION_AND_TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.3 | 10.0 | REACTOME_ACTIVATION_OF_THE_PRE_REPLICATIVE_COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.2 | 7.9 | REACTOME_DOWNREGULATION_OF_SMAD2_3_SMAD4_TRANSCRIPTIONAL_ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.2 | 4.6 | REACTOME_PYRUVATE_METABOLISM | Genes involved in Pyruvate metabolism |
| 0.2 | 5.4 | REACTOME_DEADENYLATION_OF_MRNA | Genes involved in Deadenylation of mRNA |
| 0.2 | 1.0 | REACTOME_RECYCLING_PATHWAY_OF_L1 | Genes involved in Recycling pathway of L1 |
| 0.2 | 4.2 | REACTOME_FORMATION_OF_TUBULIN_FOLDING_INTERMEDIATES_BY_CCT_TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.2 | 2.1 | REACTOME_SYNTHESIS_OF_PE | Genes involved in Synthesis of PE |
| 0.2 | 9.1 | REACTOME_ANTIVIRAL_MECHANISM_BY_IFN_STIMULATED_GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.2 | 2.7 | REACTOME_TRAF6_MEDIATED_IRF7_ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.1 | 9.3 | REACTOME_LOSS_OF_NLP_FROM_MITOTIC_CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 7.7 | REACTOME_APC_C_CDH1_MEDIATED_DEGRADATION_OF_CDC20_AND_OTHER_APC_C_CDH1_TARGETED_PROTEINS_IN_LATE_MITOSIS_EARLY_G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.1 | 7.7 | REACTOME_AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 4.4 | REACTOME_TRANSPORT_OF_MATURE_TRANSCRIPT_TO_CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.1 | 11.4 | REACTOME_MRNA_SPLICING | Genes involved in mRNA Splicing |
| 0.1 | 0.9 | REACTOME_INTRINSIC_PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 0.5 | REACTOME_ABACAVIR_TRANSPORT_AND_METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 2.2 | REACTOME_SULFUR_AMINO_ACID_METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 2.4 | REACTOME_NUCLEAR_EVENTS_KINASE_AND_TRANSCRIPTION_FACTOR_ACTIVATION | Genes involved in Nuclear Events (kinase and transcription factor activation) |
| 0.1 | 1.9 | REACTOME_SYNTHESIS_AND_INTERCONVERSION_OF_NUCLEOTIDE_DI_AND_TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 1.2 | REACTOME_PURINE_SALVAGE | Genes involved in Purine salvage |
| 0.1 | 0.8 | REACTOME_NEGATIVE_REGULATION_OF_THE_PI3K_AKT_NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 1.1 | REACTOME_EXTENSION_OF_TELOMERES | Genes involved in Extension of Telomeres |
| 0.1 | 3.7 | REACTOME_NUCLEAR_RECEPTOR_TRANSCRIPTION_PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 1.2 | REACTOME_CASPASE_MEDIATED_CLEAVAGE_OF_CYTOSKELETAL_PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 0.5 | REACTOME_ACTIVATION_OF_ATR_IN_RESPONSE_TO_REPLICATION_STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.0 | 1.0 | REACTOME_PYRIMIDINE_METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 1.2 | REACTOME_ENDOSOMAL_SORTING_COMPLEX_REQUIRED_FOR_TRANSPORT_ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.5 | REACTOME_PREFOLDIN_MEDIATED_TRANSFER_OF_SUBSTRATE_TO_CCT_TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 1.3 | REACTOME_MEIOTIC_SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 3.0 | REACTOME_ASPARAGINE_N_LINKED_GLYCOSYLATION | Genes involved in Asparagine N-linked glycosylation |
| 0.0 | 2.2 | REACTOME_GENERIC_TRANSCRIPTION_PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.0 | 1.4 | REACTOME_NCAM1_INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.6 | REACTOME_SIGNALING_BY_HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.3 | REACTOME_ACYL_CHAIN_REMODELLING_OF_PC | Genes involved in Acyl chain remodelling of PC |
| 0.0 | 1.7 | REACTOME_CLASS_B_2_SECRETIN_FAMILY_RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 1.1 | REACTOME_MITOCHONDRIAL_PROTEIN_IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 2.0 | REACTOME_PEPTIDE_CHAIN_ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.7 | REACTOME_LYSOSOME_VESICLE_BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.3 | REACTOME_SYNTHESIS_SECRETION_AND_INACTIVATION_OF_GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 0.3 | REACTOME_ANTIGEN_ACTIVATES_B_CELL_RECEPTOR_LEADING_TO_GENERATION_OF_SECOND_MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.1 | REACTOME_GAP_JUNCTION_ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.7 | REACTOME_METABOLISM_OF_VITAMINS_AND_COFACTORS | Genes involved in Metabolism of vitamins and cofactors |