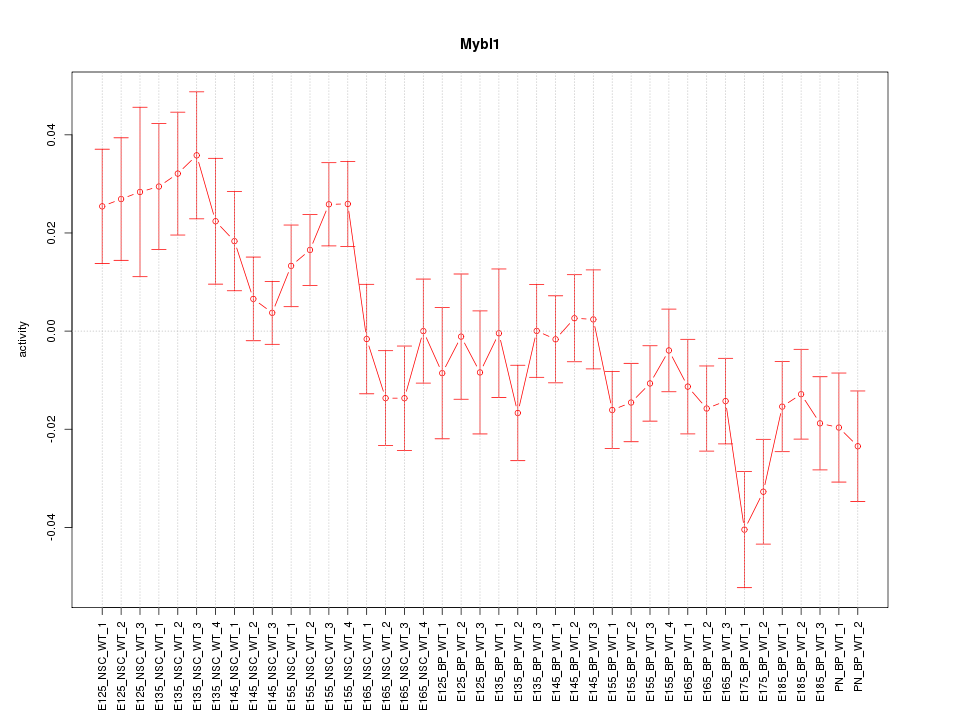
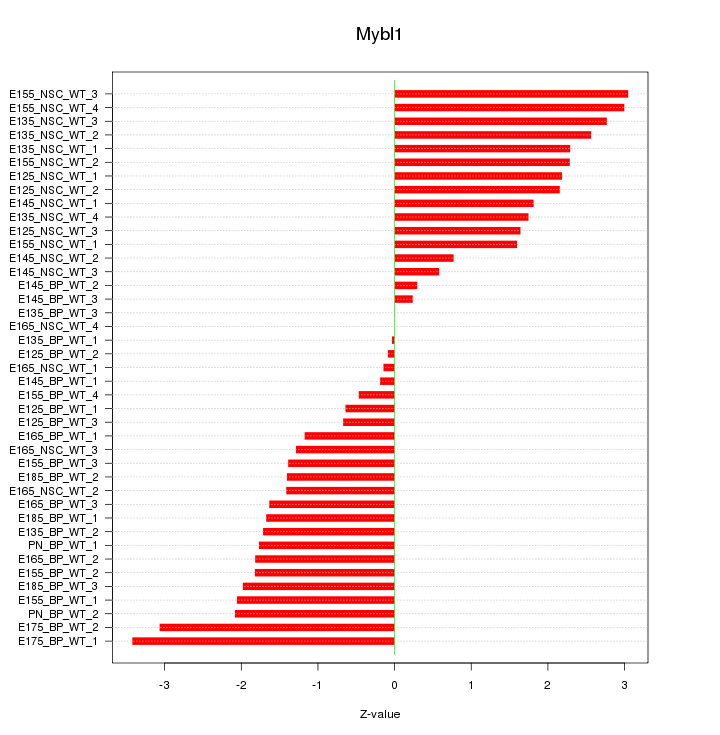
Motif ID: Mybl1
Z-value: 1.759
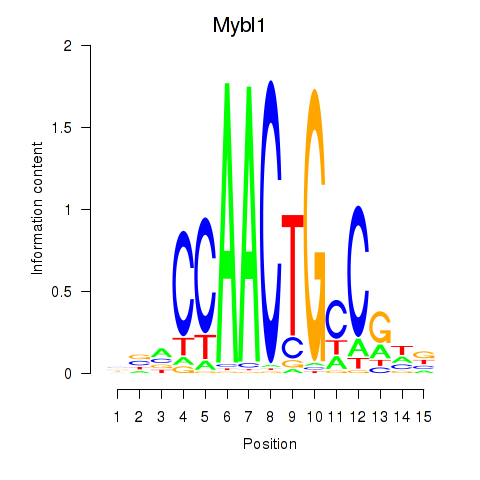
Transcription factors associated with Mybl1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Mybl1 | ENSMUSG00000025912.10 | Mybl1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Mybl1 | mm10_v2_chr1_-_9700209_9700329 | 0.27 | 8.3e-02 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.7 | 21.9 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 2.5 | 12.5 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 2.5 | 9.8 | GO:0003360 | brainstem development(GO:0003360) |
| 2.3 | 16.4 | GO:0010273 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 1.6 | 4.8 | GO:0009174 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 1.5 | 4.6 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 1.5 | 26.5 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 1.4 | 4.2 | GO:0044778 | meiotic DNA integrity checkpoint(GO:0044778) |
| 1.3 | 3.9 | GO:0051105 | regulation of DNA ligation(GO:0051105) positive regulation of DNA ligation(GO:0051106) |
| 1.2 | 6.2 | GO:0019659 | fermentation(GO:0006113) glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 1.1 | 3.4 | GO:0060535 | trachea cartilage morphogenesis(GO:0060535) |
| 1.1 | 3.4 | GO:0010814 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 1.1 | 3.3 | GO:0071649 | regulation of chemokine (C-C motif) ligand 5 production(GO:0071649) |
| 1.0 | 7.1 | GO:0000279 | M phase(GO:0000279) |
| 1.0 | 10.0 | GO:0071397 | cellular response to cholesterol(GO:0071397) |
| 1.0 | 5.0 | GO:0032901 | positive regulation of neurotrophin production(GO:0032901) |
| 1.0 | 4.0 | GO:0046552 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 1.0 | 3.0 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.9 | 7.5 | GO:0070474 | positive regulation of uterine smooth muscle contraction(GO:0070474) |
| 0.9 | 4.6 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 0.9 | 4.6 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.9 | 4.5 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.9 | 1.7 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.8 | 2.5 | GO:0045004 | DNA replication proofreading(GO:0045004) |
| 0.8 | 4.9 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.7 | 4.5 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.7 | 1.5 | GO:0060455 | negative regulation of gastric acid secretion(GO:0060455) positive regulation of somatostatin secretion(GO:0090274) |
| 0.7 | 2.2 | GO:1902162 | regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) |
| 0.7 | 2.8 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.7 | 2.1 | GO:0019405 | hexitol metabolic process(GO:0006059) alditol catabolic process(GO:0019405) fructose biosynthetic process(GO:0046370) |
| 0.7 | 16.4 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.7 | 6.7 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.7 | 9.9 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.6 | 4.5 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.6 | 1.8 | GO:0090202 | transcriptional activation by promoter-enhancer looping(GO:0071733) regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.6 | 1.7 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.6 | 1.7 | GO:1904760 | myofibroblast differentiation(GO:0036446) protein poly-ADP-ribosylation(GO:0070212) regulation of myofibroblast differentiation(GO:1904760) |
| 0.6 | 6.2 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.5 | 7.6 | GO:1900038 | negative regulation of cellular response to hypoxia(GO:1900038) |
| 0.5 | 2.7 | GO:0090666 | telomere assembly(GO:0032202) scaRNA localization to Cajal body(GO:0090666) |
| 0.5 | 2.1 | GO:0071034 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.5 | 1.6 | GO:0021577 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) lateral motor column neuron migration(GO:0097477) |
| 0.5 | 1.5 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.5 | 2.5 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.5 | 2.5 | GO:1900170 | negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.5 | 4.5 | GO:0071493 | cellular response to UV-B(GO:0071493) |
| 0.5 | 1.5 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.5 | 1.9 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.5 | 2.4 | GO:0032472 | Golgi calcium ion transport(GO:0032472) |
| 0.5 | 1.4 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.5 | 2.8 | GO:1901911 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.5 | 0.5 | GO:0061055 | myotome development(GO:0061055) |
| 0.5 | 1.8 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.4 | 3.5 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.4 | 1.7 | GO:0006244 | pyrimidine nucleotide catabolic process(GO:0006244) pyrimidine deoxyribonucleotide catabolic process(GO:0009223) |
| 0.4 | 0.9 | GO:0032714 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) negative regulation of interleukin-5 production(GO:0032714) |
| 0.4 | 1.3 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.4 | 8.4 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.4 | 12.5 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.4 | 0.4 | GO:0071609 | chemokine (C-C motif) ligand 5 production(GO:0071609) |
| 0.4 | 4.1 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.4 | 1.6 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.4 | 1.6 | GO:0030091 | protein repair(GO:0030091) |
| 0.4 | 4.5 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.4 | 0.4 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.4 | 1.1 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.4 | 2.2 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.4 | 7.9 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.4 | 2.1 | GO:0060923 | cardiac muscle cell fate commitment(GO:0060923) |
| 0.3 | 1.0 | GO:0045014 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) detection of glucose(GO:0051594) |
| 0.3 | 1.0 | GO:0002447 | eosinophil activation involved in immune response(GO:0002278) eosinophil mediated immunity(GO:0002447) eosinophil degranulation(GO:0043308) regulation of eosinophil activation(GO:1902566) |
| 0.3 | 1.3 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.3 | 2.3 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.3 | 2.6 | GO:1990173 | protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) protein localization to nucleoplasm(GO:1990173) |
| 0.3 | 1.9 | GO:0086028 | bundle of His cell to Purkinje myocyte signaling(GO:0086028) bundle of His cell action potential(GO:0086043) |
| 0.3 | 0.9 | GO:1902174 | striated muscle atrophy(GO:0014891) positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.3 | 5.0 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.3 | 0.3 | GO:0046831 | regulation of RNA export from nucleus(GO:0046831) |
| 0.3 | 5.3 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.3 | 0.9 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.3 | 1.4 | GO:0010519 | negative regulation of phospholipase activity(GO:0010519) |
| 0.3 | 1.7 | GO:0035087 | siRNA loading onto RISC involved in RNA interference(GO:0035087) |
| 0.3 | 1.7 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.3 | 1.9 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.3 | 3.6 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.3 | 1.1 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.3 | 0.8 | GO:0071707 | immunoglobulin heavy chain V-D-J recombination(GO:0071707) |
| 0.3 | 0.8 | GO:0002940 | tRNA N2-guanine methylation(GO:0002940) |
| 0.3 | 1.1 | GO:0015786 | UDP-glucose transport(GO:0015786) |
| 0.3 | 2.9 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.3 | 4.2 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.3 | 0.8 | GO:1901526 | positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.3 | 1.8 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.2 | 1.0 | GO:0019323 | pentose catabolic process(GO:0019323) |
| 0.2 | 3.6 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.2 | 1.2 | GO:1902031 | regulation of NADP metabolic process(GO:1902031) |
| 0.2 | 1.2 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.2 | 0.2 | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.2 | 1.8 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.2 | 1.6 | GO:0035701 | hematopoietic stem cell migration(GO:0035701) |
| 0.2 | 0.9 | GO:0010993 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 0.2 | 0.7 | GO:0070543 | response to linoleic acid(GO:0070543) |
| 0.2 | 4.1 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.2 | 0.9 | GO:0061188 | regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.2 | 3.1 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.2 | 0.7 | GO:0046077 | dUDP biosynthetic process(GO:0006227) pyrimidine nucleoside diphosphate metabolic process(GO:0009138) pyrimidine nucleoside diphosphate biosynthetic process(GO:0009139) pyrimidine deoxyribonucleoside diphosphate metabolic process(GO:0009196) pyrimidine deoxyribonucleoside diphosphate biosynthetic process(GO:0009197) dUDP metabolic process(GO:0046077) |
| 0.2 | 0.7 | GO:0072092 | ureteric bud invasion(GO:0072092) metanephric renal vesicle formation(GO:0072093) |
| 0.2 | 0.9 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.2 | 1.5 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.2 | 8.6 | GO:0006406 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.2 | 2.6 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.2 | 2.4 | GO:0000466 | maturation of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000466) |
| 0.2 | 1.6 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.2 | 0.6 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.2 | 0.4 | GO:0046122 | purine deoxyribonucleoside metabolic process(GO:0046122) |
| 0.2 | 3.1 | GO:0045104 | intermediate filament cytoskeleton organization(GO:0045104) |
| 0.2 | 4.6 | GO:0035115 | embryonic forelimb morphogenesis(GO:0035115) |
| 0.2 | 0.6 | GO:0002842 | positive regulation of T cell mediated immune response to tumor cell(GO:0002842) protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.2 | 1.1 | GO:0009169 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.2 | 1.1 | GO:0046015 | regulation of transcription by glucose(GO:0046015) |
| 0.2 | 0.7 | GO:2001274 | negative regulation of glucose import in response to insulin stimulus(GO:2001274) |
| 0.2 | 0.9 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.2 | 1.2 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.2 | 3.2 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.2 | 0.7 | GO:0034390 | smooth muscle cell apoptotic process(GO:0034390) regulation of smooth muscle cell apoptotic process(GO:0034391) |
| 0.2 | 1.0 | GO:0019346 | homoserine metabolic process(GO:0009092) transsulfuration(GO:0019346) |
| 0.2 | 0.7 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.2 | 0.7 | GO:0033580 | protein glycosylation at cell surface(GO:0033575) protein galactosylation at cell surface(GO:0033580) protein galactosylation(GO:0042125) |
| 0.2 | 1.5 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.2 | 0.8 | GO:1902255 | positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902255) |
| 0.2 | 0.7 | GO:0010957 | negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.2 | 1.7 | GO:0046060 | dATP metabolic process(GO:0046060) |
| 0.2 | 0.5 | GO:1904751 | positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.2 | 1.3 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.2 | 0.6 | GO:0072272 | proximal/distal pattern formation involved in metanephric nephron development(GO:0072272) |
| 0.2 | 0.5 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.2 | 3.6 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.2 | 0.8 | GO:0090343 | positive regulation of cell aging(GO:0090343) positive regulation of cellular senescence(GO:2000774) |
| 0.2 | 1.2 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 0.4 | GO:0035523 | protein K29-linked deubiquitination(GO:0035523) protein K6-linked deubiquitination(GO:0044313) |
| 0.1 | 1.6 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.1 | 6.7 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.1 | 0.7 | GO:0002155 | regulation of thyroid hormone mediated signaling pathway(GO:0002155) |
| 0.1 | 2.9 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.1 | 1.0 | GO:0031118 | rRNA pseudouridine synthesis(GO:0031118) |
| 0.1 | 1.4 | GO:0061470 | interleukin-21 production(GO:0032625) T follicular helper cell differentiation(GO:0061470) interleukin-21 secretion(GO:0072619) response to ionomycin(GO:1904636) cellular response to ionomycin(GO:1904637) |
| 0.1 | 0.8 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.1 | 0.4 | GO:1902044 | regulation of Fas signaling pathway(GO:1902044) negative regulation of Fas signaling pathway(GO:1902045) |
| 0.1 | 3.8 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 0.5 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.1 | 7.2 | GO:0070303 | negative regulation of stress-activated MAPK cascade(GO:0032873) negative regulation of stress-activated protein kinase signaling cascade(GO:0070303) |
| 0.1 | 1.3 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.1 | 2.5 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.1 | 0.6 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.1 | 0.2 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.1 | 0.5 | GO:0009597 | detection of virus(GO:0009597) |
| 0.1 | 2.3 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.1 | 4.1 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.1 | 1.0 | GO:1902969 | mitotic DNA replication(GO:1902969) |
| 0.1 | 0.3 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.1 | 3.0 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 0.6 | GO:0060484 | lung-associated mesenchyme development(GO:0060484) |
| 0.1 | 0.4 | GO:1900533 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.1 | 0.8 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.1 | 1.1 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 1.0 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.1 | 4.7 | GO:0042632 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.1 | 0.3 | GO:1901896 | positive regulation of calcium-transporting ATPase activity(GO:1901896) |
| 0.1 | 0.3 | GO:0048289 | isotype switching to IgE isotypes(GO:0048289) regulation of isotype switching to IgE isotypes(GO:0048293) |
| 0.1 | 0.4 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.1 | 1.2 | GO:0001711 | endodermal cell fate commitment(GO:0001711) |
| 0.1 | 2.0 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 0.7 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.1 | 2.2 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.1 | 1.6 | GO:0016445 | somatic diversification of immunoglobulins(GO:0016445) |
| 0.1 | 0.3 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.1 | 0.3 | GO:0006183 | GTP biosynthetic process(GO:0006183) |
| 0.1 | 1.4 | GO:0001553 | luteinization(GO:0001553) |
| 0.1 | 0.6 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.1 | 1.2 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.1 | 1.0 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.1 | 5.7 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.1 | 0.9 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.1 | 1.2 | GO:0010883 | regulation of lipid storage(GO:0010883) |
| 0.1 | 0.6 | GO:0032693 | negative regulation of interleukin-10 production(GO:0032693) |
| 0.1 | 0.4 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) |
| 0.1 | 3.0 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.1 | 1.1 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.1 | 2.0 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 0.7 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 | 0.1 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.1 | 0.9 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.1 | 2.1 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.1 | 0.5 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.1 | 0.5 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.1 | 0.2 | GO:0052428 | modulation of molecular function in other organism(GO:0044359) negative regulation of molecular function in other organism(GO:0044362) negative regulation of molecular function in other organism involved in symbiotic interaction(GO:0052204) modulation of molecular function in other organism involved in symbiotic interaction(GO:0052205) negative regulation by host of symbiont molecular function(GO:0052405) modification by host of symbiont molecular function(GO:0052428) |
| 0.1 | 1.4 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.1 | 2.4 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 0.9 | GO:0010763 | positive regulation of fibroblast migration(GO:0010763) |
| 0.1 | 4.9 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.1 | 1.0 | GO:0048712 | negative regulation of astrocyte differentiation(GO:0048712) |
| 0.1 | 0.1 | GO:2000504 | positive regulation of blood vessel remodeling(GO:2000504) |
| 0.1 | 0.7 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.1 | 0.7 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.1 | 0.6 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.1 | 0.7 | GO:0046929 | negative regulation of neurotransmitter secretion(GO:0046929) |
| 0.1 | 1.1 | GO:1901984 | negative regulation of protein acetylation(GO:1901984) negative regulation of peptidyl-lysine acetylation(GO:2000757) |
| 0.1 | 0.9 | GO:0039703 | viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.0 | 0.8 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 1.4 | GO:0003016 | respiratory system process(GO:0003016) |
| 0.0 | 0.7 | GO:0043278 | response to isoquinoline alkaloid(GO:0014072) response to morphine(GO:0043278) |
| 0.0 | 7.0 | GO:0000082 | G1/S transition of mitotic cell cycle(GO:0000082) |
| 0.0 | 0.3 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 0.8 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 0.9 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.0 | 0.9 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 1.1 | GO:1902042 | negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.0 | 0.3 | GO:0042492 | gamma-delta T cell differentiation(GO:0042492) |
| 0.0 | 0.3 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.0 | 0.5 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.4 | GO:0048146 | positive regulation of fibroblast proliferation(GO:0048146) |
| 0.0 | 1.6 | GO:0048663 | neuron fate commitment(GO:0048663) |
| 0.0 | 0.7 | GO:0033108 | mitochondrial respiratory chain complex assembly(GO:0033108) |
| 0.0 | 0.8 | GO:0000956 | nuclear-transcribed mRNA catabolic process(GO:0000956) |
| 0.0 | 0.5 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.8 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 | 0.2 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) regulation of parathyroid hormone secretion(GO:2000828) |
| 0.0 | 0.6 | GO:0001702 | gastrulation with mouth forming second(GO:0001702) |
| 0.0 | 0.9 | GO:0071514 | genetic imprinting(GO:0071514) |
| 0.0 | 0.4 | GO:0097194 | execution phase of apoptosis(GO:0097194) |
| 0.0 | 0.2 | GO:0010917 | negative regulation of mitochondrial membrane potential(GO:0010917) |
| 0.0 | 0.2 | GO:0018208 | peptidyl-proline modification(GO:0018208) |
| 0.0 | 1.5 | GO:0008584 | male gonad development(GO:0008584) development of primary male sexual characteristics(GO:0046546) |
| 0.0 | 0.3 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.2 | GO:0032782 | bile acid secretion(GO:0032782) |
| 0.0 | 2.4 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.3 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) aggresome assembly(GO:0070842) |
| 0.0 | 1.7 | GO:0008637 | apoptotic mitochondrial changes(GO:0008637) |
| 0.0 | 0.5 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.9 | GO:0046785 | microtubule polymerization(GO:0046785) |
| 0.0 | 0.2 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 1.0 | GO:0051384 | response to glucocorticoid(GO:0051384) |
| 0.0 | 0.2 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 0.9 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 1.1 | GO:1900181 | negative regulation of protein localization to nucleus(GO:1900181) |
| 0.0 | 0.2 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.0 | 1.4 | GO:0030177 | positive regulation of Wnt signaling pathway(GO:0030177) |
| 0.0 | 1.9 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 0.5 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.0 | 0.8 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.2 | GO:0017144 | drug metabolic process(GO:0017144) |
| 0.0 | 0.3 | GO:0072337 | modified amino acid transport(GO:0072337) |
| 0.0 | 0.2 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 0.8 | GO:0045727 | positive regulation of translation(GO:0045727) |
| 0.0 | 0.6 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 1.3 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.0 | 0.1 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 0.5 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.2 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.0 | 0.9 | GO:0070507 | regulation of microtubule cytoskeleton organization(GO:0070507) |
| 0.0 | 0.3 | GO:0006801 | superoxide metabolic process(GO:0006801) |
| 0.0 | 0.1 | GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) |
| 0.0 | 0.2 | GO:0034063 | stress granule assembly(GO:0034063) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 9.8 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
| 2.0 | 21.7 | GO:0005642 | annulate lamellae(GO:0005642) |
| 1.6 | 4.7 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 1.3 | 4.0 | GO:0044299 | C-fiber(GO:0044299) |
| 1.3 | 3.8 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 1.0 | 3.0 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.8 | 31.0 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.8 | 3.3 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.8 | 3.2 | GO:0045251 | mitochondrial electron transfer flavoprotein complex(GO:0017133) electron transfer flavoprotein complex(GO:0045251) |
| 0.7 | 4.4 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.7 | 4.2 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.7 | 3.4 | GO:0031523 | Myb complex(GO:0031523) |
| 0.7 | 3.4 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.6 | 3.6 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.6 | 2.2 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.6 | 15.5 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.5 | 1.6 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.5 | 1.6 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.5 | 2.5 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.5 | 3.0 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.5 | 2.0 | GO:0043293 | apoptosome(GO:0043293) |
| 0.5 | 3.5 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.4 | 11.1 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.4 | 2.5 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.4 | 1.9 | GO:0099524 | region of cytosol(GO:0099522) postsynaptic cytosol(GO:0099524) |
| 0.4 | 1.1 | GO:1990047 | spindle matrix(GO:1990047) |
| 0.3 | 1.7 | GO:0097226 | sperm mitochondrial sheath(GO:0097226) |
| 0.3 | 1.0 | GO:0000811 | GINS complex(GO:0000811) |
| 0.3 | 4.5 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.3 | 4.7 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.3 | 2.2 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.3 | 2.9 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.3 | 1.9 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.3 | 1.1 | GO:0045283 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.3 | 4.6 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.3 | 1.3 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.3 | 1.8 | GO:0060293 | P granule(GO:0043186) pole plasm(GO:0045495) germ plasm(GO:0060293) |
| 0.2 | 2.7 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.2 | 2.4 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.2 | 2.6 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.2 | 5.8 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.2 | 2.5 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.2 | 1.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.2 | 17.3 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.2 | 1.5 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.2 | 5.2 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.2 | 1.3 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.2 | 2.1 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.2 | 3.0 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.2 | 0.9 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.2 | 4.0 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.2 | 5.7 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.2 | 1.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.2 | 0.9 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.2 | 1.9 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.2 | 2.8 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.2 | 10.1 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.2 | 0.5 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.2 | 0.8 | GO:0072487 | MSL complex(GO:0072487) |
| 0.2 | 1.2 | GO:0034709 | methylosome(GO:0034709) |
| 0.1 | 1.0 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.1 | 4.8 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.1 | 0.3 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 1.0 | GO:0097546 | ciliary base(GO:0097546) |
| 0.1 | 1.6 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.1 | 14.9 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 4.2 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 1.2 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.1 | 1.7 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 9.1 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 1.2 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 0.9 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 12.0 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 1.4 | GO:0034518 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.1 | 0.7 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 0.9 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.1 | 7.0 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 0.9 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 1.6 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.1 | 0.6 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 1.3 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 0.9 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.1 | 0.7 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 0.3 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 1.1 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 4.0 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 0.8 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) |
| 0.1 | 1.8 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.1 | 1.1 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 1.0 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.1 | 6.6 | GO:0000776 | kinetochore(GO:0000776) |
| 0.1 | 0.5 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 1.3 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.1 | 2.5 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.1 | 0.3 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.1 | 1.7 | GO:0030496 | midbody(GO:0030496) |
| 0.1 | 12.2 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.1 | 1.8 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.1 | 0.2 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.1 | 0.5 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.1 | 4.3 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 1.5 | GO:0098839 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.1 | 0.6 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 3.8 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.1 | 4.4 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 0.9 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.7 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.0 | 0.7 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.1 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.0 | 0.1 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.0 | 1.6 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.0 | 2.6 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.1 | GO:0097651 | phosphatidylinositol 3-kinase complex, class I(GO:0097651) |
| 0.0 | 1.2 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.8 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.9 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 6.1 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.9 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.9 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 1.3 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.9 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.3 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 3.5 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 3.9 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 2.2 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.5 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.0 | 1.4 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.0 | 0.7 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.4 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.2 | GO:0005922 | connexon complex(GO:0005922) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 12.5 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 1.6 | 9.8 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 1.6 | 7.9 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 1.6 | 4.7 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 1.5 | 4.6 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 1.4 | 21.8 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 1.2 | 7.1 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 1.2 | 5.9 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 1.2 | 3.5 | GO:0071207 | histone pre-mRNA stem-loop binding(GO:0071207) |
| 1.1 | 4.6 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 1.1 | 6.7 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 1.1 | 4.5 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.8 | 3.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.8 | 2.3 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.8 | 4.5 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.7 | 2.2 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.6 | 2.5 | GO:0038049 | glucocorticoid receptor activity(GO:0004883) transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.6 | 5.0 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.6 | 6.7 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.6 | 3.0 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.6 | 4.1 | GO:1901612 | phosphatidylglycerol binding(GO:1901611) cardiolipin binding(GO:1901612) |
| 0.6 | 3.9 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.5 | 15.9 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.5 | 2.5 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.5 | 4.5 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.5 | 2.8 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.5 | 1.4 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.5 | 2.8 | GO:0000298 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.4 | 2.1 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.4 | 2.8 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.4 | 1.2 | GO:0004473 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.4 | 6.3 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.4 | 4.4 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.4 | 2.9 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.4 | 2.5 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.4 | 1.8 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.3 | 8.1 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.3 | 5.5 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.3 | 2.2 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.3 | 1.2 | GO:1990269 | RNA polymerase II C-terminal domain phosphoserine binding(GO:1990269) |
| 0.3 | 0.9 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.3 | 1.7 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.3 | 1.1 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.3 | 1.6 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.3 | 1.0 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.2 | 4.6 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.2 | 0.7 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.2 | 0.7 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.2 | 1.5 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.2 | 1.9 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.2 | 2.1 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.2 | 1.7 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.2 | 0.6 | GO:0003979 | UDP-glucose 6-dehydrogenase activity(GO:0003979) |
| 0.2 | 1.7 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.2 | 1.0 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.2 | 0.6 | GO:0071936 | coreceptor activity involved in Wnt signaling pathway(GO:0071936) |
| 0.2 | 1.9 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) G-rich strand telomeric DNA binding(GO:0098505) N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.2 | 3.4 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.2 | 3.8 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.2 | 2.8 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.2 | 1.1 | GO:0046404 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.2 | 1.3 | GO:0015288 | porin activity(GO:0015288) |
| 0.2 | 2.7 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.2 | 0.8 | GO:0031543 | peptidyl-proline dioxygenase activity(GO:0031543) |
| 0.2 | 0.6 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.2 | 7.6 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.2 | 0.8 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.2 | 0.6 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.2 | 0.5 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.2 | 0.8 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.1 | 0.9 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.1 | 0.7 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) |
| 0.1 | 1.8 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 10.7 | GO:0004004 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.1 | 0.9 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 1.6 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.1 | 4.3 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 0.5 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.1 | 0.6 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.1 | 2.2 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 1.0 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.1 | 0.7 | GO:0047276 | N-acetyllactosaminide 3-alpha-galactosyltransferase activity(GO:0047276) |
| 0.1 | 1.3 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.1 | 0.3 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.1 | 3.0 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 2.4 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.1 | 10.4 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.1 | 3.3 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.1 | 7.3 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.1 | 0.7 | GO:0048101 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.1 | 0.7 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.1 | 0.6 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 0.4 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.1 | 3.3 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 2.6 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.1 | 1.6 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.1 | 0.9 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 0.5 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.1 | 0.4 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.1 | 3.2 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.1 | 0.3 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.1 | 3.8 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 5.4 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 13.7 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 3.3 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 0.3 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.1 | 0.9 | GO:0048038 | quinone binding(GO:0048038) |
| 0.1 | 0.2 | GO:0008297 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) |
| 0.1 | 1.3 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.1 | 3.2 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.1 | 3.2 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 0.7 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 8.0 | GO:0008201 | heparin binding(GO:0008201) |
| 0.1 | 3.0 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 3.0 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.1 | 3.1 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.1 | 1.0 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.1 | 5.2 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 0.4 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 1.0 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 1.0 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 1.3 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 1.2 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 1.0 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 1.8 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 4.4 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.1 | 1.1 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.1 | 2.6 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.1 | 2.9 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 1.4 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.7 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.4 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.5 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.2 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 1.5 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.7 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 1.8 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.8 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.8 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 4.6 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.5 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.1 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.0 | 0.3 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.4 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.3 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 2.0 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 0.2 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.0 | 0.5 | GO:0015923 | mannosidase activity(GO:0015923) |
| 0.0 | 1.6 | GO:0051287 | NAD binding(GO:0051287) |
| 0.0 | 0.4 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 1.0 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.2 | GO:0051429 | corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.0 | 1.4 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.6 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.0 | 1.1 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.5 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.3 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 1.1 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 1.0 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.5 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.6 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.3 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.3 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 1.5 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.1 | GO:0044388 | small protein activating enzyme binding(GO:0044388) |
| 0.0 | 0.7 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 0.2 | GO:0051861 | glycolipid binding(GO:0051861) |
| 0.0 | 0.1 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.2 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.0 | 0.0 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.5 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.2 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 4.0 | GO:0003682 | chromatin binding(GO:0003682) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 15.6 | PID_SMAD2_3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.5 | 4.7 | SA_G2_AND_M_PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.4 | 4.6 | PID_TCR_JNK_PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.3 | 10.2 | PID_EPHRINB_REV_PATHWAY | Ephrin B reverse signaling |
| 0.2 | 14.6 | PID_PLK1_PATHWAY | PLK1 signaling events |
| 0.2 | 8.3 | PID_HNF3B_PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 2.0 | PID_ERB_GENOMIC_PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 4.0 | PID_TRAIL_PATHWAY | TRAIL signaling pathway |
| 0.1 | 3.4 | PID_HDAC_CLASSIII_PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 1.1 | PID_ALK2_PATHWAY | ALK2 signaling events |
| 0.1 | 4.6 | PID_AURORA_B_PATHWAY | Aurora B signaling |
| 0.1 | 3.3 | SA_B_CELL_RECEPTOR_COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.1 | 3.6 | PID_ATR_PATHWAY | ATR signaling pathway |
| 0.1 | 1.7 | SA_CASPASE_CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 4.5 | PID_HIF1_TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 3.6 | PID_E2F_PATHWAY | E2F transcription factor network |
| 0.1 | 3.4 | PID_SMAD2_3NUCLEAR_PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.1 | 4.2 | PID_TELOMERASE_PATHWAY | Regulation of Telomerase |
| 0.1 | 0.9 | PID_AVB3_OPN_PATHWAY | Osteopontin-mediated events |
| 0.0 | 0.9 | PID_IL2_STAT5_PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.6 | PID_HIF1A_PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 1.3 | PID_BMP_PATHWAY | BMP receptor signaling |
| 0.0 | 1.9 | ST_T_CELL_SIGNAL_TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 2.0 | PID_REG_GR_PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 1.7 | PID_FOXO_PATHWAY | FoxO family signaling |
| 0.0 | 0.4 | PID_AURORA_A_PATHWAY | Aurora A signaling |
| 0.0 | 1.1 | PID_LIS1_PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 1.4 | PID_P75_NTR_PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.6 | PID_TOLL_ENDOGENOUS_PATHWAY | Endogenous TLR signaling |
| 0.0 | 1.2 | PID_P53_REGULATION_PATHWAY | p53 pathway |
| 0.0 | 1.2 | PID_LKB1_PATHWAY | LKB1 signaling events |
| 0.0 | 0.9 | PID_TGFBR_PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.6 | NABA_PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.8 | PID_AR_PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.1 | PID_RANBP2_PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.3 | ST_INTERLEUKIN_4_PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 0.2 | PID_PRL_SIGNALING_EVENTS_PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.2 | PID_AMB2_NEUTROPHILS_PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.6 | ST_INTEGRIN_SIGNALING_PATHWAY | Integrin Signaling Pathway |
| 0.0 | 1.0 | WNT_SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 1.5 | NABA_ECM_GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.4 | PID_ILK_PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.2 | PID_PI3KCI_PATHWAY | Class I PI3K signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 4.6 | REACTOME_ETHANOL_OXIDATION | Genes involved in Ethanol oxidation |
| 1.0 | 5.9 | REACTOME_TRANSLOCATION_OF_ZAP_70_TO_IMMUNOLOGICAL_SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.8 | 21.9 | REACTOME_PTM_GAMMA_CARBOXYLATION_HYPUSINE_FORMATION_AND_ARYLSULFATASE_ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.6 | 9.9 | REACTOME_ASSOCIATION_OF_LICENSING_FACTORS_WITH_THE_PRE_REPLICATIVE_COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.6 | 14.9 | REACTOME_KINESINS | Genes involved in Kinesins |
| 0.4 | 7.1 | REACTOME_CONVERSION_FROM_APC_C_CDC20_TO_APC_C_CDH1_IN_LATE_ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.3 | 8.2 | REACTOME_GLUTATHIONE_CONJUGATION | Genes involved in Glutathione conjugation |
| 0.3 | 3.5 | REACTOME_SLBP_DEPENDENT_PROCESSING_OF_REPLICATION_DEPENDENT_HISTONE_PRE_MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.3 | 4.0 | REACTOME_HOMOLOGOUS_RECOMBINATION_REPAIR_OF_REPLICATION_INDEPENDENT_DOUBLE_STRAND_BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.3 | 8.3 | REACTOME_EXTENSION_OF_TELOMERES | Genes involved in Extension of Telomeres |
| 0.3 | 4.9 | REACTOME_TRAF6_MEDIATED_IRF7_ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.3 | 5.9 | REACTOME_FORMATION_OF_TUBULIN_FOLDING_INTERMEDIATES_BY_CCT_TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.3 | 6.8 | REACTOME_PRE_NOTCH_TRANSCRIPTION_AND_TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.2 | 12.9 | REACTOME_AMYLOIDS | Genes involved in Amyloids |
| 0.2 | 4.8 | REACTOME_PYRIMIDINE_METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.2 | 2.8 | REACTOME_HDL_MEDIATED_LIPID_TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.2 | 2.1 | REACTOME_MRNA_DECAY_BY_3_TO_5_EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.2 | 21.5 | REACTOME_MITOTIC_PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.2 | 3.3 | REACTOME_REGULATION_OF_INSULIN_LIKE_GROWTH_FACTOR_IGF_ACTIVITY_BY_INSULIN_LIKE_GROWTH_FACTOR_BINDING_PROTEINS_IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.2 | 14.1 | REACTOME_ANTIVIRAL_MECHANISM_BY_IFN_STIMULATED_GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.2 | 4.5 | REACTOME_PYRUVATE_METABOLISM | Genes involved in Pyruvate metabolism |
| 0.2 | 5.4 | REACTOME_G1_PHASE | Genes involved in G1 Phase |
| 0.2 | 4.6 | REACTOME_SYNTHESIS_AND_INTERCONVERSION_OF_NUCLEOTIDE_DI_AND_TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.2 | 3.4 | REACTOME_G0_AND_EARLY_G1 | Genes involved in G0 and Early G1 |
| 0.2 | 5.1 | REACTOME_DOWNREGULATION_OF_SMAD2_3_SMAD4_TRANSCRIPTIONAL_ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.1 | 2.8 | REACTOME_AMINE_COMPOUND_SLC_TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.1 | 1.8 | REACTOME_PROCESSING_OF_INTRONLESS_PRE_MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.1 | 1.2 | REACTOME_CTNNB1_PHOSPHORYLATION_CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.1 | 6.0 | REACTOME_FORMATION_OF_THE_TERNARY_COMPLEX_AND_SUBSEQUENTLY_THE_43S_COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 7.7 | REACTOME_RESPIRATORY_ELECTRON_TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 2.0 | REACTOME_DEADENYLATION_OF_MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 1.2 | REACTOME_DESTABILIZATION_OF_MRNA_BY_KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.1 | 1.2 | REACTOME_PREFOLDIN_MEDIATED_TRANSFER_OF_SUBSTRATE_TO_CCT_TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.1 | 1.5 | REACTOME_PURINE_SALVAGE | Genes involved in Purine salvage |
| 0.1 | 1.8 | REACTOME_RNA_POL_III_CHAIN_ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.1 | 0.7 | REACTOME_INTRINSIC_PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 2.1 | REACTOME_CYTOSOLIC_TRNA_AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 3.6 | REACTOME_AUTODEGRADATION_OF_CDH1_BY_CDH1_APC_C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.1 | 0.6 | REACTOME_GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 4.3 | REACTOME_3_UTR_MEDIATED_TRANSLATIONAL_REGULATION | Genes involved in 3' -UTR-mediated translational regulation |
| 0.1 | 0.8 | REACTOME_NONSENSE_MEDIATED_DECAY_ENHANCED_BY_THE_EXON_JUNCTION_COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.1 | 1.6 | REACTOME_RNA_POL_II_TRANSCRIPTION_PRE_INITIATION_AND_PROMOTER_OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.1 | 1.0 | REACTOME_UNWINDING_OF_DNA | Genes involved in Unwinding of DNA |
| 0.1 | 1.6 | REACTOME_CLEAVAGE_OF_GROWING_TRANSCRIPT_IN_THE_TERMINATION_REGION_ | Genes involved in Cleavage of Growing Transcript in the Termination Region |
| 0.1 | 1.3 | REACTOME_SIGNALING_BY_BMP | Genes involved in Signaling by BMP |
| 0.1 | 1.1 | REACTOME_TGF_BETA_RECEPTOR_SIGNALING_IN_EMT_EPITHELIAL_TO_MESENCHYMAL_TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 2.7 | REACTOME_TRANSCRIPTIONAL_REGULATION_OF_WHITE_ADIPOCYTE_DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 3.4 | REACTOME_CIRCADIAN_CLOCK | Genes involved in Circadian Clock |
| 0.1 | 0.6 | REACTOME_HYALURONAN_UPTAKE_AND_DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 0.5 | REACTOME_SYNTHESIS_SECRETION_AND_INACTIVATION_OF_GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 0.9 | REACTOME_NEGATIVE_REGULATORS_OF_RIG_I_MDA5_SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.3 | REACTOME_DOWNSTREAM_SIGNAL_TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 1.0 | REACTOME_REGULATION_OF_GENE_EXPRESSION_IN_BETA_CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.1 | REACTOME_G_BETA_GAMMA_SIGNALLING_THROUGH_PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 2.5 | REACTOME_ACTIVATION_OF_CHAPERONE_GENES_BY_XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.6 | REACTOME_OXYGEN_DEPENDENT_PROLINE_HYDROXYLATION_OF_HYPOXIA_INDUCIBLE_FACTOR_ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.4 | REACTOME_RNA_POL_III_TRANSCRIPTION_INITIATION_FROM_TYPE_3_PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 3.5 | REACTOME_MRNA_SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.3 | REACTOME_LIPOPROTEIN_METABOLISM | Genes involved in Lipoprotein metabolism |
| 0.0 | 1.6 | REACTOME_GLUCAGON_TYPE_LIGAND_RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 6.3 | REACTOME_ANTIGEN_PROCESSING_UBIQUITINATION_PROTEASOME_DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 2.7 | REACTOME_PEPTIDE_LIGAND_BINDING_RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.3 | REACTOME_MTORC1_MEDIATED_SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.7 | REACTOME_ASSOCIATION_OF_TRIC_CCT_WITH_TARGET_PROTEINS_DURING_BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 1.1 | REACTOME_CLASS_B_2_SECRETIN_FAMILY_RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 1.4 | REACTOME_MITOCHONDRIAL_PROTEIN_IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.3 | REACTOME_PURINE_RIBONUCLEOSIDE_MONOPHOSPHATE_BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.8 | REACTOME_HS_GAG_BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 2.0 | REACTOME_SIGNALING_BY_RHO_GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.4 | REACTOME_SYNTHESIS_OF_PC | Genes involved in Synthesis of PC |
| 0.0 | 0.7 | REACTOME_O_LINKED_GLYCOSYLATION_OF_MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 1.5 | REACTOME_CELL_DEATH_SIGNALLING_VIA_NRAGE_NRIF_AND_NADE | Genes involved in Cell death signalling via NRAGE, NRIF and NADE |
| 0.0 | 0.3 | REACTOME_INSULIN_RECEPTOR_RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.2 | REACTOME_GAP_JUNCTION_ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 1.1 | REACTOME_GLYCEROPHOSPHOLIPID_BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 0.3 | REACTOME_GAP_JUNCTION_TRAFFICKING | Genes involved in Gap junction trafficking |
| 0.0 | 0.1 | REACTOME_REGULATION_OF_IFNG_SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.2 | REACTOME_ACTIVATED_AMPK_STIMULATES_FATTY_ACID_OXIDATION_IN_MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.2 | REACTOME_TIE2_SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.2 | REACTOME_ALPHA_LINOLENIC_ACID_ALA_METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |