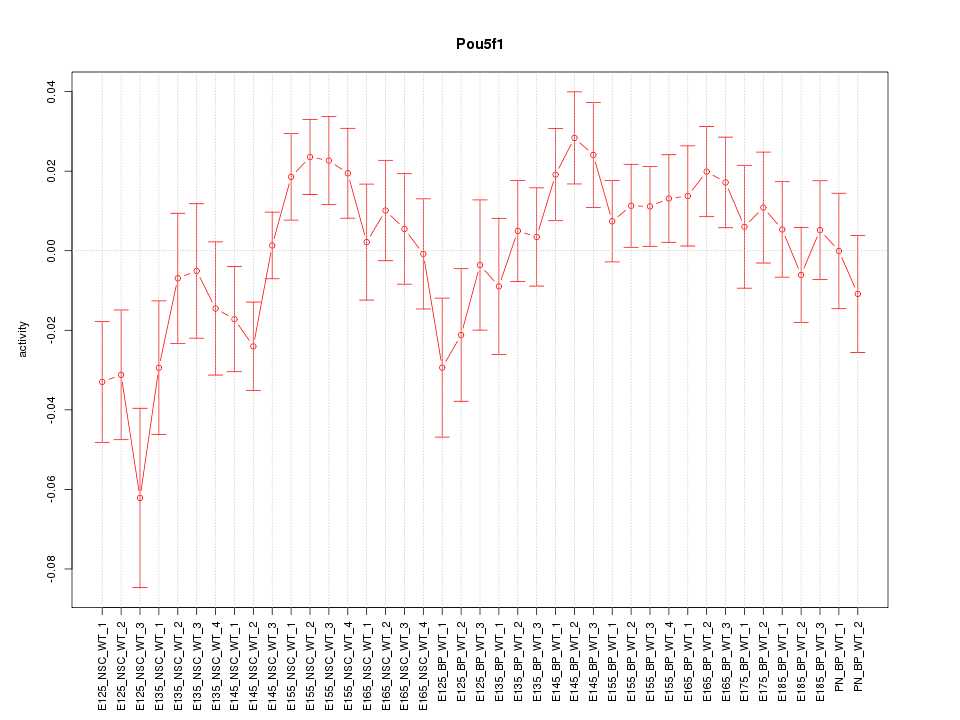
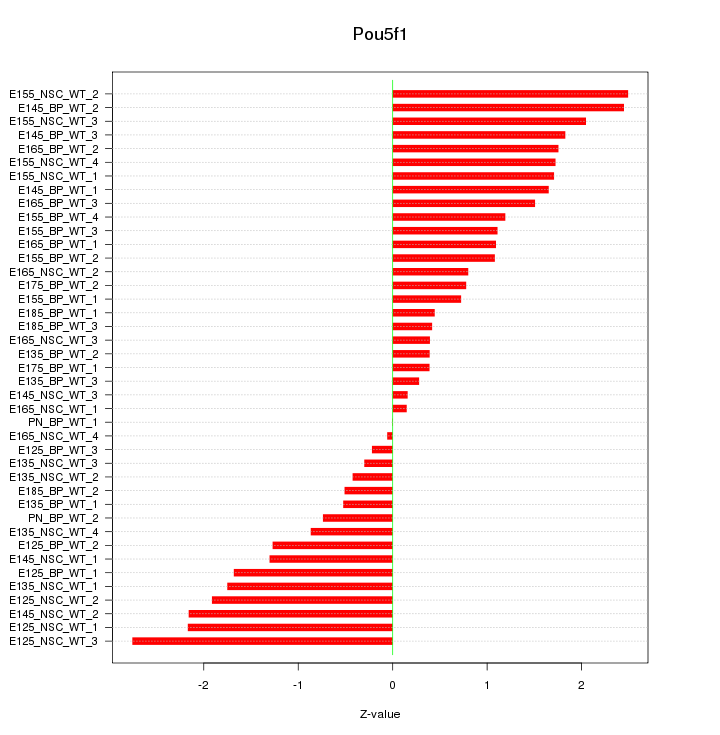
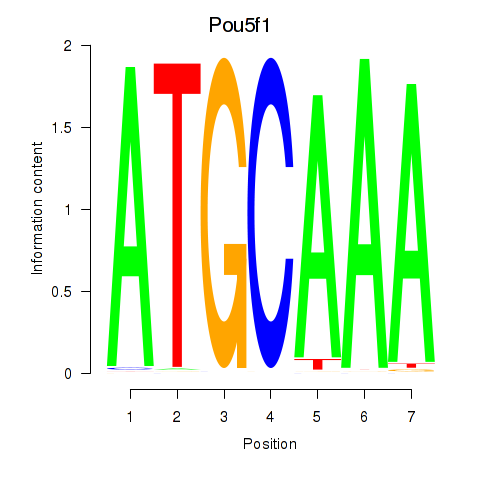
Motif ID: Pou5f1
Z-value: 1.339
Transcription factors associated with Pou5f1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Pou5f1 | ENSMUSG00000024406.10 | Pou5f1 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 8.8 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 2.4 | 9.6 | GO:1905167 | positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 1.7 | 5.0 | GO:0048687 | positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) |
| 1.6 | 6.5 | GO:1903288 | positive regulation of potassium ion import(GO:1903288) |
| 1.6 | 4.8 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 1.2 | 3.6 | GO:1902938 | regulation of intracellular calcium activated chloride channel activity(GO:1902938) |
| 1.2 | 4.7 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 1.1 | 4.4 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 1.0 | 3.8 | GO:1902340 | telomeric heterochromatin assembly(GO:0031509) negative regulation of chromosome condensation(GO:1902340) |
| 1.0 | 13.3 | GO:0045161 | neuronal ion channel clustering(GO:0045161) |
| 0.9 | 3.7 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.8 | 4.2 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) |
| 0.8 | 1.7 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.8 | 3.4 | GO:0090126 | protein complex assembly involved in synapse maturation(GO:0090126) |
| 0.8 | 3.4 | GO:2000110 | protein sialylation(GO:1990743) negative regulation of macrophage apoptotic process(GO:2000110) |
| 0.8 | 11.5 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.8 | 8.8 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.8 | 6.9 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.8 | 2.3 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.7 | 2.9 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.7 | 2.2 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.7 | 4.2 | GO:0032308 | positive regulation of prostaglandin secretion(GO:0032308) |
| 0.7 | 2.1 | GO:0010871 | negative regulation of receptor biosynthetic process(GO:0010871) |
| 0.7 | 4.1 | GO:0042045 | epithelial fluid transport(GO:0042045) |
| 0.7 | 2.0 | GO:0031554 | regulation of DNA-templated transcription, termination(GO:0031554) |
| 0.7 | 8.7 | GO:0072189 | ureter development(GO:0072189) |
| 0.7 | 4.0 | GO:2001012 | mesenchymal cell differentiation involved in kidney development(GO:0072161) mesenchymal cell differentiation involved in renal system development(GO:2001012) |
| 0.6 | 5.8 | GO:0042756 | drinking behavior(GO:0042756) |
| 0.6 | 1.9 | GO:0002182 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.6 | 8.0 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.6 | 3.1 | GO:0055095 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) |
| 0.6 | 9.4 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.6 | 8.1 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.6 | 3.4 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.6 | 7.4 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.6 | 1.7 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.5 | 2.7 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.5 | 1.6 | GO:0003289 | atrial septum primum morphogenesis(GO:0003289) |
| 0.5 | 1.4 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.4 | 2.6 | GO:0097491 | trigeminal nerve morphogenesis(GO:0021636) trigeminal nerve structural organization(GO:0021637) trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) sympathetic neuron projection extension(GO:0097490) sympathetic neuron projection guidance(GO:0097491) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.4 | 2.2 | GO:1901843 | positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.4 | 1.3 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.4 | 2.0 | GO:0042636 | negative regulation of hair cycle(GO:0042636) progesterone secretion(GO:0042701) regulation of follicle-stimulating hormone secretion(GO:0046880) follicle-stimulating hormone secretion(GO:0046884) negative regulation of hair follicle development(GO:0051799) positive regulation of ovulation(GO:0060279) |
| 0.4 | 2.4 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.4 | 1.2 | GO:1901228 | positive regulation of transcription from RNA polymerase II promoter involved in heart development(GO:1901228) |
| 0.4 | 2.7 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.4 | 2.7 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.4 | 2.3 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) cellular response to zinc ion(GO:0071294) |
| 0.4 | 13.0 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.4 | 1.1 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) positive regulation of NLRP3 inflammasome complex assembly(GO:1900227) |
| 0.4 | 1.9 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.3 | 2.3 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.3 | 1.6 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.3 | 16.0 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.3 | 2.0 | GO:0033139 | regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033139) |
| 0.3 | 1.9 | GO:1903056 | positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.3 | 1.5 | GO:2001015 | skeletal muscle satellite cell activation(GO:0014719) negative regulation of skeletal muscle cell differentiation(GO:2001015) |
| 0.3 | 1.8 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.3 | 0.9 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 0.3 | 1.4 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.3 | 0.6 | GO:0061324 | limb joint morphogenesis(GO:0036022) embryonic skeletal limb joint morphogenesis(GO:0036023) positive regulation of epithelial cell proliferation involved in prostate gland development(GO:0060769) canonical Wnt signaling pathway involved in positive regulation of cardiac outflow tract cell proliferation(GO:0061324) regulation of centromeric sister chromatid cohesion(GO:0070602) regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901963) |
| 0.3 | 8.9 | GO:0030325 | adrenal gland development(GO:0030325) |
| 0.3 | 1.1 | GO:2000481 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) positive regulation of cAMP-dependent protein kinase activity(GO:2000481) |
| 0.3 | 2.4 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.3 | 2.4 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.3 | 1.0 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) negative regulation of cholesterol efflux(GO:0090370) |
| 0.3 | 2.4 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.2 | 3.5 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) |
| 0.2 | 3.9 | GO:0048520 | positive regulation of behavior(GO:0048520) |
| 0.2 | 2.7 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.2 | 1.1 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.2 | 1.6 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.2 | 0.9 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.2 | 16.1 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.2 | 2.3 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) |
| 0.2 | 11.4 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.2 | 2.1 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.2 | 2.2 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.2 | 2.9 | GO:0002230 | positive regulation of defense response to virus by host(GO:0002230) |
| 0.2 | 1.6 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.2 | 4.7 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.1 | 0.4 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.1 | 1.3 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 1.6 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.1 | 0.7 | GO:0070494 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.1 | 5.5 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 0.8 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 0.5 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.1 | 0.4 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 0.1 | 0.6 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.1 | 0.7 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.1 | 20.7 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.1 | 1.3 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.1 | 3.4 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 0.7 | GO:2000645 | negative regulation of receptor catabolic process(GO:2000645) |
| 0.1 | 0.6 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.1 | 0.4 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.1 | 0.7 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.1 | 2.0 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.1 | 4.2 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.1 | 1.4 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.1 | 1.0 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.1 | 1.8 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.1 | 0.6 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.1 | 2.7 | GO:0001541 | ovarian follicle development(GO:0001541) |
| 0.1 | 1.6 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.1 | 1.1 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.1 | 0.3 | GO:0010804 | negative regulation of tumor necrosis factor-mediated signaling pathway(GO:0010804) |
| 0.1 | 1.7 | GO:1900078 | positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.1 | 1.0 | GO:0001539 | cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.1 | 1.7 | GO:0010762 | regulation of fibroblast migration(GO:0010762) |
| 0.0 | 3.1 | GO:0032755 | positive regulation of interleukin-6 production(GO:0032755) |
| 0.0 | 0.6 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.2 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.0 | 5.2 | GO:0098656 | anion transmembrane transport(GO:0098656) |
| 0.0 | 0.7 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.4 | GO:0031111 | negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 0.0 | 0.2 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 3.4 | GO:0000045 | autophagosome assembly(GO:0000045) |
| 0.0 | 0.7 | GO:0090083 | regulation of inclusion body assembly(GO:0090083) |
| 0.0 | 1.2 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.6 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.0 | 2.3 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 4.0 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.6 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.4 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.0 | 1.1 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.5 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 1.4 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 3.8 | GO:0048813 | dendrite morphogenesis(GO:0048813) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.3 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.0 | 0.5 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.6 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.0 | 2.8 | GO:0051056 | regulation of small GTPase mediated signal transduction(GO:0051056) |
| 0.0 | 0.2 | GO:0043388 | positive regulation of DNA binding(GO:0043388) |
| 0.0 | 0.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.5 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 2.3 | GO:0006402 | mRNA catabolic process(GO:0006402) |
| 0.0 | 0.5 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.7 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.3 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 1.0 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.0 | 0.1 | GO:1900194 | receptor guanylyl cyclase signaling pathway(GO:0007168) negative regulation of oocyte development(GO:0060283) negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.1 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 9.7 | GO:0044308 | axonal spine(GO:0044308) |
| 1.4 | 4.1 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.9 | 6.5 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.8 | 12.8 | GO:0098839 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.7 | 13.9 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.7 | 2.0 | GO:0043512 | inhibin A complex(GO:0043512) |
| 0.5 | 2.7 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.5 | 1.6 | GO:1903095 | microprocessor complex(GO:0070877) ribonuclease III complex(GO:1903095) |
| 0.5 | 4.8 | GO:0000805 | X chromosome(GO:0000805) |
| 0.5 | 6.9 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.4 | 3.4 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.4 | 4.7 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.4 | 8.0 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.3 | 3.4 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.2 | 2.1 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.2 | 3.7 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.2 | 6.2 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.2 | 9.6 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.2 | 6.3 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 14.1 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 1.7 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 2.0 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 0.7 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 1.0 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 2.4 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 0.6 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.1 | 0.4 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.1 | 0.7 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 1.3 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.1 | 1.3 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 1.5 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 1.9 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 0.3 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.1 | 6.7 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 11.6 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 0.4 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.1 | 1.7 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 1.1 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 1.6 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.1 | 1.7 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 3.8 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 3.6 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 1.1 | GO:0034704 | calcium channel complex(GO:0034704) |
| 0.1 | 7.4 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.1 | 2.3 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 11.0 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.1 | 1.9 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 1.5 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 6.4 | GO:0034702 | ion channel complex(GO:0034702) |
| 0.0 | 0.9 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 3.4 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 2.0 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 6.3 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 2.0 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 8.9 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.0 | 1.7 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.8 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 2.0 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.0 | 0.5 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 3.8 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 1.5 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.0 | 0.9 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 2.6 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 2.0 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.5 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.6 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 2.0 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.9 | GO:0005814 | centriole(GO:0005814) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 9.6 | GO:0016964 | alpha-2 macroglobulin receptor activity(GO:0016964) |
| 2.9 | 8.8 | GO:0002153 | steroid receptor RNA activator RNA binding(GO:0002153) |
| 2.3 | 16.0 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 2.2 | 8.8 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 1.4 | 4.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 1.2 | 8.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 1.1 | 3.4 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 1.1 | 6.5 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 1.0 | 5.1 | GO:0055100 | adiponectin binding(GO:0055100) |
| 1.0 | 5.0 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.8 | 3.4 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.8 | 2.3 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.7 | 2.2 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.7 | 3.4 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.7 | 2.0 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.6 | 6.5 | GO:0008556 | sodium:potassium-exchanging ATPase activity(GO:0005391) potassium-transporting ATPase activity(GO:0008556) |
| 0.6 | 3.7 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.6 | 13.3 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.5 | 2.7 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.5 | 7.9 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.5 | 2.3 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.4 | 5.6 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.4 | 3.8 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.4 | 6.9 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.3 | 2.0 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.3 | 6.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.3 | 1.6 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.3 | 4.2 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.3 | 11.1 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.3 | 2.1 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.2 | 3.4 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.2 | 1.5 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) axon guidance receptor activity(GO:0008046) |
| 0.2 | 3.4 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.2 | 0.7 | GO:0016901 | glycerol-3-phosphate dehydrogenase activity(GO:0004368) oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 0.2 | 2.7 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.2 | 4.2 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.2 | 1.4 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 0.2 | 1.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.2 | 0.7 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 0.2 | 4.2 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.2 | 1.4 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.2 | 1.0 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.2 | 1.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.2 | 4.4 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.2 | 4.7 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.1 | 4.0 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.1 | 2.7 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 0.8 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.1 | 0.6 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.1 | 0.7 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.1 | 0.4 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.1 | 8.6 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.1 | 1.9 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 9.7 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.1 | 2.2 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 2.0 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 5.5 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 8.0 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.1 | 0.8 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 1.6 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 5.5 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.1 | 0.5 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 0.3 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.1 | 1.9 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 0.7 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 2.1 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 7.7 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 1.6 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 5.1 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 3.2 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.1 | 2.9 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.1 | 0.6 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.1 | 1.8 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 0.4 | GO:0034979 | NAD-dependent protein deacetylase activity(GO:0034979) |
| 0.1 | 0.2 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.0 | 0.6 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.2 | GO:1990188 | euchromatin binding(GO:1990188) |
| 0.0 | 3.3 | GO:0019209 | kinase activator activity(GO:0019209) |
| 0.0 | 1.7 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.0 | 13.8 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.3 | GO:0030553 | cGMP binding(GO:0030553) 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 1.2 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 3.9 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.0 | 2.1 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 2.2 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.3 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 9.3 | GO:0016887 | ATPase activity(GO:0016887) |
| 0.0 | 0.4 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.4 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.6 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 2.1 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.0 | 0.0 | GO:0019002 | GMP binding(GO:0019002) |
| 0.0 | 5.1 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 4.7 | GO:0008289 | lipid binding(GO:0008289) |
| 0.0 | 0.5 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 16.3 | PID_HNF3A_PATHWAY | FOXA1 transcription factor network |
| 0.3 | 9.6 | PID_UPA_UPAR_PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.3 | 10.9 | PID_RXR_VDR_PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.2 | 4.2 | PID_ANTHRAX_PATHWAY | Cellular roles of Anthrax toxin |
| 0.2 | 4.2 | PID_CIRCADIAN_PATHWAY | Circadian rhythm pathway |
| 0.2 | 4.4 | ST_WNT_CA2_CYCLIC_GMP_PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.2 | 6.9 | PID_NCADHERIN_PATHWAY | N-cadherin signaling events |
| 0.2 | 10.5 | PID_EPHB_FWD_PATHWAY | EPHB forward signaling |
| 0.2 | 3.7 | PID_INTEGRIN_A9B1_PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.2 | 8.0 | PID_ATR_PATHWAY | ATR signaling pathway |
| 0.2 | 2.3 | PID_ERBB_NETWORK_PATHWAY | ErbB receptor signaling network |
| 0.1 | 6.3 | PID_SHP2_PATHWAY | SHP2 signaling |
| 0.1 | 2.3 | ST_ADRENERGIC | Adrenergic Pathway |
| 0.1 | 8.6 | PID_REG_GR_PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 3.9 | ST_TUMOR_NECROSIS_FACTOR_PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.1 | 12.5 | PID_PDGFRB_PATHWAY | PDGFR-beta signaling pathway |
| 0.1 | 2.9 | PID_REELIN_PATHWAY | Reelin signaling pathway |
| 0.1 | 1.0 | SA_CASPASE_CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 0.5 | PID_AVB3_INTEGRIN_PATHWAY | Integrins in angiogenesis |
| 0.1 | 7.3 | NABA_ECM_AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 4.1 | PID_HDAC_CLASSI_PATHWAY | Signaling events mediated by HDAC Class I |
| 0.1 | 2.0 | PID_ALK1_PATHWAY | ALK1 signaling events |
| 0.1 | 0.5 | ST_GRANULE_CELL_SURVIVAL_PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.1 | 1.8 | PID_CD8_TCR_PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.1 | 2.7 | PID_TGFBR_PATHWAY | TGF-beta receptor signaling |
| 0.1 | 2.4 | PID_AR_TF_PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.7 | PID_TCPTP_PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 2.1 | PID_LKB1_PATHWAY | LKB1 signaling events |
| 0.0 | 0.9 | PID_ALPHA_SYNUCLEIN_PATHWAY | Alpha-synuclein signaling |
| 0.0 | 4.2 | NABA_ECM_REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.1 | PID_AR_PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.5 | PID_HEDGEHOG_GLI_PATHWAY | Hedgehog signaling events mediated by Gli proteins |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 18.9 | REACTOME_UNBLOCKING_OF_NMDA_RECEPTOR_GLUTAMATE_BINDING_AND_ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.5 | 2.0 | REACTOME_GLYCOPROTEIN_HORMONES | Genes involved in Glycoprotein hormones |
| 0.5 | 6.8 | REACTOME_DEGRADATION_OF_THE_EXTRACELLULAR_MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.4 | 14.9 | REACTOME_MYOGENESIS | Genes involved in Myogenesis |
| 0.3 | 3.4 | REACTOME_TERMINATION_OF_O_GLYCAN_BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.3 | 2.8 | REACTOME_RNA_POL_III_TRANSCRIPTION_INITIATION_FROM_TYPE_3_PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.2 | 1.4 | REACTOME_ELEVATION_OF_CYTOSOLIC_CA2_LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.2 | 6.6 | REACTOME_ADHERENS_JUNCTIONS_INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.2 | 4.2 | REACTOME_CRMPS_IN_SEMA3A_SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.2 | 4.7 | REACTOME_OTHER_SEMAPHORIN_INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.2 | 6.1 | REACTOME_BASIGIN_INTERACTIONS | Genes involved in Basigin interactions |
| 0.2 | 1.6 | REACTOME_PYRIMIDINE_CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.2 | 4.7 | REACTOME_RNA_POL_III_TRANSCRIPTION_TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.2 | 1.7 | REACTOME_ACETYLCHOLINE_NEUROTRANSMITTER_RELEASE_CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.1 | 2.7 | REACTOME_HORMONE_SENSITIVE_LIPASE_HSL_MEDIATED_TRIACYLGLYCEROL_HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 9.6 | REACTOME_NUCLEAR_RECEPTOR_TRANSCRIPTION_PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 2.3 | REACTOME_LIGAND_GATED_ION_CHANNEL_TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.1 | 2.7 | REACTOME_SYNTHESIS_OF_PC | Genes involved in Synthesis of PC |
| 0.1 | 2.7 | REACTOME_NEPHRIN_INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 1.6 | REACTOME_MICRORNA_MIRNA_BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.1 | 2.6 | REACTOME_DARPP_32_EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 1.3 | REACTOME_METABOLISM_OF_STEROID_HORMONES_AND_VITAMINS_A_AND_D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.1 | 2.0 | REACTOME_BMAL1_CLOCK_NPAS2_ACTIVATES_CIRCADIAN_EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 0.3 | REACTOME_PLATELET_ADHESION_TO_EXPOSED_COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 2.0 | REACTOME_NOTCH1_INTRACELLULAR_DOMAIN_REGULATES_TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 2.3 | REACTOME_INHIBITION_OF_VOLTAGE_GATED_CA2_CHANNELS_VIA_GBETA_GAMMA_SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 4.0 | REACTOME_NRAGE_SIGNALS_DEATH_THROUGH_JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.5 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_GOLGI_MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 1.8 | REACTOME_NCAM1_INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 3.7 | REACTOME_INTEGRIN_CELL_SURFACE_INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.7 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_EARLY_ENDOSOME_MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.6 | REACTOME_PURINE_RIBONUCLEOSIDE_MONOPHOSPHATE_BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 1.0 | REACTOME_FATTY_ACYL_COA_BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.0 | 0.4 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_PLASMA_MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.6 | REACTOME_VOLTAGE_GATED_POTASSIUM_CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.9 | REACTOME_LOSS_OF_NLP_FROM_MITOTIC_CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.8 | REACTOME_GENERIC_TRANSCRIPTION_PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.0 | 1.1 | REACTOME_FACTORS_INVOLVED_IN_MEGAKARYOCYTE_DEVELOPMENT_AND_PLATELET_PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |