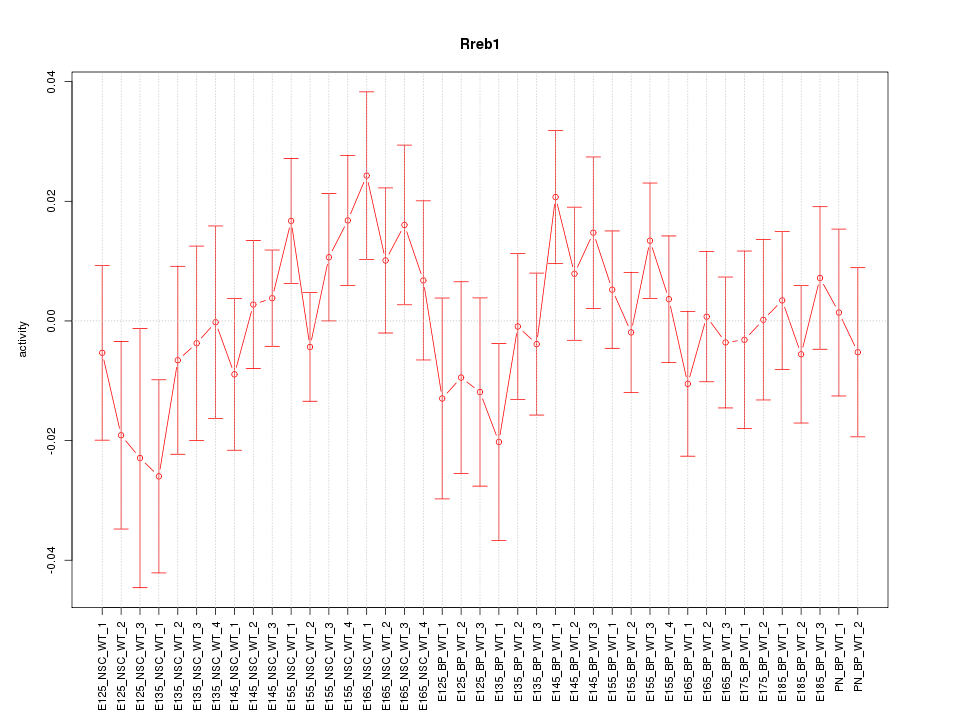
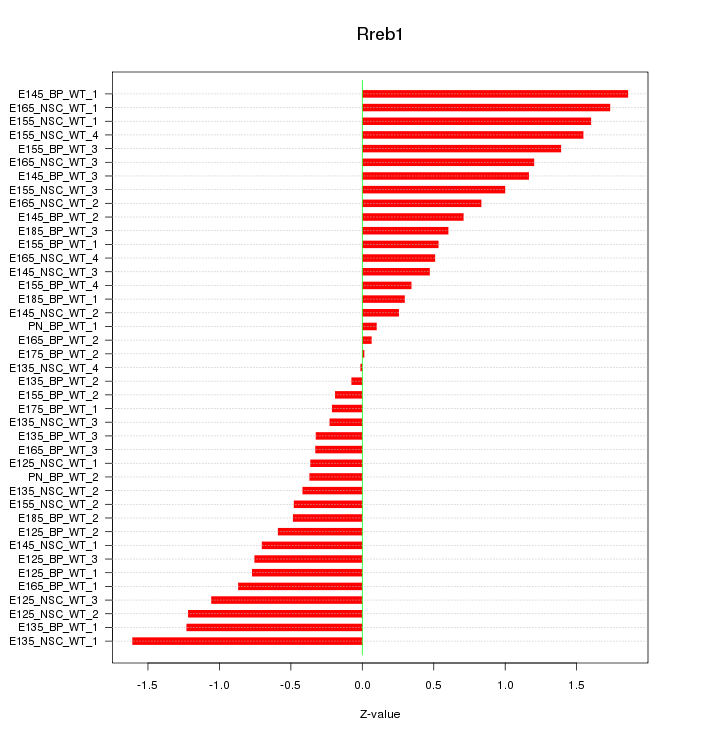
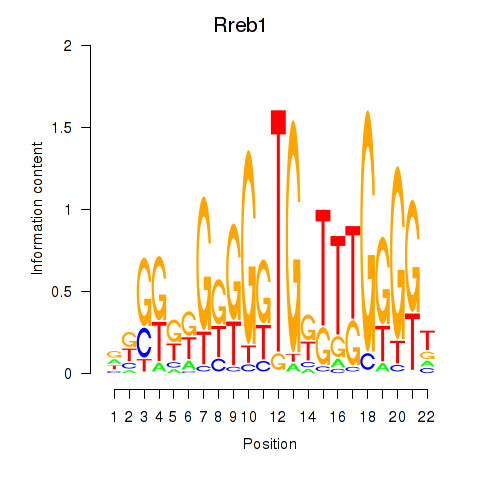
Motif ID: Rreb1
Z-value: 0.863
Transcription factors associated with Rreb1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Rreb1 | ENSMUSG00000039087.10 | Rreb1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Rreb1 | mm10_v2_chr13_+_37826225_37826315 | 0.03 | 8.6e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 4.8 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.9 | 2.8 | GO:0030827 | negative regulation of cGMP metabolic process(GO:0030824) negative regulation of cGMP biosynthetic process(GO:0030827) negative regulation of guanylate cyclase activity(GO:0031283) |
| 0.9 | 2.6 | GO:1904049 | negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 0.6 | 3.8 | GO:1903056 | positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.5 | 1.4 | GO:0048687 | positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) |
| 0.4 | 2.2 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.4 | 2.1 | GO:0034436 | glycoprotein transport(GO:0034436) |
| 0.4 | 1.7 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.4 | 1.6 | GO:0010986 | positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.4 | 2.8 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.3 | 1.0 | GO:1902528 | regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.3 | 1.6 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.3 | 1.0 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.3 | 0.9 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.3 | 0.9 | GO:0048007 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.3 | 7.1 | GO:0095500 | acetylcholine receptor signaling pathway(GO:0095500) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.3 | 0.8 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.3 | 2.1 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.3 | 1.3 | GO:0070460 | thyroid-stimulating hormone secretion(GO:0070460) |
| 0.2 | 1.0 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.2 | 0.9 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.2 | 2.2 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) |
| 0.2 | 1.1 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.2 | 2.6 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.2 | 1.6 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.2 | 0.6 | GO:1903011 | negative regulation of bone development(GO:1903011) |
| 0.2 | 1.9 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 0.2 | 3.8 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.2 | 2.8 | GO:0043045 | DNA methylation involved in embryo development(GO:0043045) changes to DNA methylation involved in embryo development(GO:1901538) |
| 0.2 | 0.7 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.2 | 0.9 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.1 | 0.3 | GO:0045963 | negative regulation of catecholamine metabolic process(GO:0045914) negative regulation of dopamine metabolic process(GO:0045963) |
| 0.1 | 1.9 | GO:0014052 | regulation of gamma-aminobutyric acid secretion(GO:0014052) |
| 0.1 | 0.6 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.1 | 0.5 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.1 | 0.4 | GO:0045359 | positive regulation of interferon-beta biosynthetic process(GO:0045359) |
| 0.1 | 0.8 | GO:0061622 | glycolytic process through glucose-1-phosphate(GO:0061622) |
| 0.1 | 0.4 | GO:0046077 | dUDP biosynthetic process(GO:0006227) pyrimidine nucleoside diphosphate metabolic process(GO:0009138) pyrimidine nucleoside diphosphate biosynthetic process(GO:0009139) deoxyribonucleoside diphosphate biosynthetic process(GO:0009189) pyrimidine deoxyribonucleoside diphosphate metabolic process(GO:0009196) pyrimidine deoxyribonucleoside diphosphate biosynthetic process(GO:0009197) dUDP metabolic process(GO:0046077) |
| 0.1 | 0.3 | GO:0051884 | regulation of anagen(GO:0051884) |
| 0.1 | 0.6 | GO:0038032 | termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.1 | 0.4 | GO:0042939 | glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.1 | 0.9 | GO:0070494 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.1 | 0.4 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.1 | 0.7 | GO:0048597 | post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.1 | 0.5 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.1 | 1.0 | GO:0071397 | cellular response to cholesterol(GO:0071397) |
| 0.1 | 0.6 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.1 | 0.7 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.1 | 0.3 | GO:0014891 | striated muscle atrophy(GO:0014891) |
| 0.1 | 0.7 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 0.3 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 0.1 | 0.8 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.1 | 0.6 | GO:0051790 | short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.1 | 2.0 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.1 | 0.2 | GO:0010726 | positive regulation of hydrogen peroxide metabolic process(GO:0010726) |
| 0.1 | 1.1 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.1 | 0.3 | GO:0018103 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.1 | 0.3 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.1 | 0.3 | GO:0002554 | serotonin secretion by platelet(GO:0002554) interleukin-3 production(GO:0032632) beta selection(GO:0043366) |
| 0.1 | 0.4 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.1 | 1.0 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 1.3 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.1 | 0.3 | GO:0051342 | regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.1 | 0.7 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.1 | 2.2 | GO:0032933 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.1 | 2.7 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.1 | 0.8 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 1.7 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.1 | 0.4 | GO:0034441 | plasma lipoprotein particle oxidation(GO:0034441) |
| 0.1 | 0.4 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 0.1 | 0.3 | GO:0060468 | prevention of polyspermy(GO:0060468) |
| 0.1 | 0.8 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 0.6 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 0.5 | GO:0042748 | circadian sleep/wake cycle, non-REM sleep(GO:0042748) |
| 0.1 | 0.5 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.1 | 0.2 | GO:0045869 | negative regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045869) |
| 0.1 | 1.1 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.1 | 0.2 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.1 | 0.2 | GO:0015920 | lipopolysaccharide transport(GO:0015920) |
| 0.1 | 4.2 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 | 0.3 | GO:1903028 | regulation of opsonization(GO:1903027) positive regulation of opsonization(GO:1903028) |
| 0.1 | 3.7 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.1 | 0.9 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.1 | 1.1 | GO:0060081 | membrane hyperpolarization(GO:0060081) |
| 0.1 | 0.7 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.1 | 0.3 | GO:0090315 | negative regulation of protein targeting to membrane(GO:0090315) |
| 0.1 | 0.5 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.1 | 0.3 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.1 | 0.7 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 1.2 | GO:0042092 | type 2 immune response(GO:0042092) |
| 0.1 | 1.3 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.1 | 0.6 | GO:0060644 | mammary gland epithelial cell differentiation(GO:0060644) |
| 0.1 | 2.4 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.1 | 0.6 | GO:0061470 | interleukin-21 production(GO:0032625) T follicular helper cell differentiation(GO:0061470) interleukin-21 secretion(GO:0072619) |
| 0.1 | 0.4 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 0.6 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.1 | 0.3 | GO:0032329 | serine transport(GO:0032329) |
| 0.1 | 0.6 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.1 | 0.6 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.1 | 0.1 | GO:0090649 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.1 | 0.6 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.1 | 1.9 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.1 | 0.2 | GO:0048239 | negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 0.1 | 0.1 | GO:1900133 | renin secretion into blood stream(GO:0002001) regulation of renin secretion into blood stream(GO:1900133) |
| 0.0 | 0.7 | GO:0003283 | atrial septum development(GO:0003283) |
| 0.0 | 0.4 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.3 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.7 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.2 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.0 | 8.2 | GO:0051017 | actin filament bundle assembly(GO:0051017) actin filament bundle organization(GO:0061572) |
| 0.0 | 0.4 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.6 | GO:0030828 | positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.0 | 0.4 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 1.3 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.2 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.0 | 4.5 | GO:0007098 | centrosome cycle(GO:0007098) |
| 0.0 | 1.2 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.0 | 0.1 | GO:0046618 | urate transport(GO:0015747) drug export(GO:0046618) negative regulation of intestinal absorption(GO:1904479) |
| 0.0 | 0.5 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.7 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.8 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.0 | 0.2 | GO:1902856 | negative regulation of nonmotile primary cilium assembly(GO:1902856) |
| 0.0 | 0.3 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.0 | GO:0072069 | DCT cell differentiation(GO:0072069) metanephric DCT cell differentiation(GO:0072240) |
| 0.0 | 0.1 | GO:1900095 | regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.0 | 0.4 | GO:0010954 | positive regulation of protein processing(GO:0010954) |
| 0.0 | 0.2 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.0 | 0.2 | GO:0098909 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) regulation of cardiac muscle cell action potential involved in regulation of contraction(GO:0098909) |
| 0.0 | 0.1 | GO:0071267 | amino acid salvage(GO:0043102) L-methionine salvage(GO:0071267) |
| 0.0 | 1.3 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 | 0.3 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.2 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.2 | GO:0035428 | hexose transmembrane transport(GO:0035428) |
| 0.0 | 0.2 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.1 | GO:0002940 | tRNA N2-guanine methylation(GO:0002940) |
| 0.0 | 0.3 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.0 | 0.6 | GO:0046039 | GTP metabolic process(GO:0046039) |
| 0.0 | 0.4 | GO:0032469 | endoplasmic reticulum calcium ion homeostasis(GO:0032469) |
| 0.0 | 0.3 | GO:0072348 | sulfur compound transport(GO:0072348) |
| 0.0 | 0.2 | GO:1901032 | negative regulation of response to reactive oxygen species(GO:1901032) negative regulation of hydrogen peroxide-induced cell death(GO:1903206) |
| 0.0 | 0.1 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 0.4 | GO:0051926 | negative regulation of calcium ion transport(GO:0051926) |
| 0.0 | 0.3 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.0 | 0.6 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 0.1 | GO:0033089 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.0 | 0.6 | GO:0001541 | ovarian follicle development(GO:0001541) |
| 0.0 | 0.4 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 0.3 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.1 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.4 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.4 | GO:1901379 | regulation of potassium ion transmembrane transport(GO:1901379) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.5 | GO:0046579 | positive regulation of Ras protein signal transduction(GO:0046579) |
| 0.0 | 1.1 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.2 | GO:0030934 | collagen type XII trimer(GO:0005595) anchoring collagen complex(GO:0030934) |
| 1.0 | 3.8 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.5 | 2.6 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.3 | 7.3 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.2 | 2.8 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.2 | 0.8 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.2 | 1.0 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.2 | 0.7 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.2 | 2.1 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.2 | 0.9 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.2 | 2.2 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 0.7 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 1.1 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.1 | 0.9 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.1 | 1.0 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.1 | 4.2 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.1 | 1.3 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 0.7 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.1 | 0.4 | GO:1990462 | omegasome(GO:1990462) |
| 0.1 | 0.4 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.1 | 1.6 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.1 | 1.3 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 1.1 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 0.9 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.1 | 1.5 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 0.3 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.1 | 0.4 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.1 | 1.6 | GO:0009986 | cell surface(GO:0009986) |
| 0.1 | 0.3 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 0.9 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 0.7 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.1 | 0.8 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.4 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.5 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 1.9 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.6 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.7 | GO:0044754 | autolysosome(GO:0044754) |
| 0.0 | 1.0 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 1.1 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.8 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 0.5 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.6 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 1.0 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 1.5 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 1.0 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 5.0 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.3 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.5 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.7 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.6 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 1.3 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.2 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.2 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.2 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 1.0 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.6 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 0.2 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 1.2 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.6 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 1.8 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 3.4 | GO:0044306 | neuron projection terminus(GO:0044306) |
| 0.0 | 0.6 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.6 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.8 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.3 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.1 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.4 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 3.1 | GO:0019898 | extrinsic component of membrane(GO:0019898) |
| 0.0 | 0.2 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 0.3 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.7 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 1.5 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 3.1 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 1.4 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 2.3 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 3.0 | GO:0000139 | Golgi membrane(GO:0000139) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 8.5 | GO:0030549 | acetylcholine receptor activator activity(GO:0030549) |
| 0.8 | 4.8 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.7 | 2.1 | GO:0034437 | very-low-density lipoprotein particle binding(GO:0034189) glycoprotein transporter activity(GO:0034437) |
| 0.7 | 2.8 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.4 | 1.6 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.3 | 1.0 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.3 | 1.6 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.3 | 1.6 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.3 | 1.2 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.3 | 1.5 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.3 | 0.9 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.3 | 2.3 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.3 | 1.4 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.3 | 0.8 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.2 | 1.0 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.2 | 0.9 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 0.2 | 0.8 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.2 | 0.8 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.2 | 0.6 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.2 | 0.6 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.2 | 3.0 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.2 | 0.7 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.2 | 2.1 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.2 | 0.8 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.2 | 0.6 | GO:0004315 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) |
| 0.2 | 4.6 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 0.4 | GO:0004127 | cytidylate kinase activity(GO:0004127) uridylate kinase activity(GO:0009041) |
| 0.1 | 0.6 | GO:1990188 | euchromatin binding(GO:1990188) |
| 0.1 | 0.7 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.1 | 0.3 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.1 | 3.5 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 4.0 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 0.7 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.1 | 0.3 | GO:0043758 | acetate-CoA ligase (ADP-forming) activity(GO:0043758) |
| 0.1 | 0.5 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.1 | 0.3 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.1 | 0.7 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 0.3 | GO:0036004 | GAF domain binding(GO:0036004) |
| 0.1 | 1.7 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 1.0 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.1 | 1.6 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 0.4 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.1 | 1.1 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.1 | 0.2 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.1 | 1.0 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.1 | 0.8 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.1 | 0.3 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.1 | 1.5 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.1 | 0.6 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.1 | 0.4 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.1 | 0.7 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.1 | 0.4 | GO:0051429 | corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.1 | 0.7 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 0.3 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.1 | 0.5 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.1 | 2.3 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 0.5 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.1 | 0.3 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.1 | 0.2 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 0.6 | GO:0019531 | secondary active sulfate transmembrane transporter activity(GO:0008271) oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 0.4 | GO:0015651 | quaternary ammonium group transmembrane transporter activity(GO:0015651) |
| 0.0 | 0.6 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.4 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.0 | 0.4 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.0 | 0.2 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 0.2 | GO:0097003 | adipokinetic hormone receptor activity(GO:0097003) |
| 0.0 | 0.4 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.0 | 2.2 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 2.4 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.4 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.3 | GO:0035325 | receptor signaling protein tyrosine kinase activity(GO:0004716) Toll-like receptor binding(GO:0035325) |
| 0.0 | 0.3 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 2.4 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 0.7 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.2 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.1 | GO:0051379 | epinephrine binding(GO:0051379) |
| 0.0 | 0.6 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.9 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 0.9 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.4 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.0 | 0.1 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.0 | 0.4 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.2 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.7 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.5 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.1 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 1.2 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 1.1 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 4.2 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.4 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.2 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 0.5 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 0.4 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 1.1 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.4 | GO:0005267 | potassium channel activity(GO:0005267) |
| 0.0 | 1.0 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 0.6 | GO:0042562 | hormone binding(GO:0042562) |
| 0.0 | 0.1 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.6 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.6 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.7 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 1.4 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 3.7 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.4 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.2 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.2 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) |
| 0.0 | 2.3 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.0 | 0.2 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.3 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.1 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.0 | 0.4 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.8 | GO:0016798 | hydrolase activity, acting on glycosyl bonds(GO:0016798) |
| 0.0 | 0.6 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.3 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.6 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 2.2 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 0.3 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.3 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 0.4 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.3 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.1 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.2 | SA_CASPASE_CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 4.1 | PID_SYNDECAN_1_PATHWAY | Syndecan-1-mediated signaling events |
| 0.1 | 2.1 | PID_UPA_UPAR_PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 0.3 | SA_FAS_SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 0.7 | PID_VEGF_VEGFR_PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 0.6 | PID_INTEGRIN5_PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 2.2 | PID_EPO_PATHWAY | EPO signaling pathway |
| 0.0 | 1.3 | PID_NFKAPPAB_CANONICAL_PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 1.5 | PID_RXR_VDR_PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.4 | PID_S1P_S1P1_PATHWAY | S1P1 pathway |
| 0.0 | 3.3 | PID_HDAC_CLASSI_PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.3 | PID_WNT_CANONICAL_PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.2 | ST_B_CELL_ANTIGEN_RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 1.4 | PID_SHP2_PATHWAY | SHP2 signaling |
| 0.0 | 0.9 | PID_ARF_3PATHWAY | Arf1 pathway |
| 0.0 | 0.3 | PID_NFKAPPAB_ATYPICAL_PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 1.1 | SIG_CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 2.1 | PID_P53_REGULATION_PATHWAY | p53 pathway |
| 0.0 | 0.7 | ST_WNT_BETA_CATENIN_PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.9 | SIG_BCR_SIGNALING_PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 0.4 | PID_TOLL_ENDOGENOUS_PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.9 | PID_CASPASE_PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.2 | PID_IL6_7_PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.7 | PID_PI3KCI_PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.4 | PID_P38_ALPHA_BETA_PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.3 | PID_FCER1_PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.4 | PID_LIS1_PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.5 | PID_CDC42_REG_PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.3 | SIG_CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 0.2 | PID_REELIN_PATHWAY | Reelin signaling pathway |
| 0.0 | 0.5 | PID_MYC_REPRESS_PATHWAY | Validated targets of C-MYC transcriptional repression |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.1 | REACTOME_EICOSANOID_LIGAND_BINDING_RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.2 | 0.9 | REACTOME_ARMS_MEDIATED_ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.1 | 1.0 | REACTOME_TANDEM_PORE_DOMAIN_POTASSIUM_CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.1 | 1.7 | REACTOME_AMINE_LIGAND_BINDING_RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 4.5 | REACTOME_ADHERENS_JUNCTIONS_INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 1.3 | REACTOME_AMINE_DERIVED_HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 2.2 | REACTOME_SIGNAL_ATTENUATION | Genes involved in Signal attenuation |
| 0.1 | 5.7 | REACTOME_COLLAGEN_FORMATION | Genes involved in Collagen formation |
| 0.1 | 1.7 | REACTOME_ALPHA_LINOLENIC_ACID_ALA_METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 0.4 | REACTOME_SYNTHESIS_SECRETION_AND_DEACYLATION_OF_GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 0.8 | REACTOME_SYNTHESIS_OF_PE | Genes involved in Synthesis of PE |
| 0.1 | 0.3 | REACTOME_ACTIVATION_OF_NF_KAPPAB_IN_B_CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.1 | 0.4 | REACTOME_ORGANIC_CATION_ANION_ZWITTERION_TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 1.0 | REACTOME_SYNTHESIS_OF_SUBSTRATES_IN_N_GLYCAN_BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 0.7 | REACTOME_VEGF_LIGAND_RECEPTOR_INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 1.9 | REACTOME_NITRIC_OXIDE_STIMULATES_GUANYLATE_CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.1 | 2.4 | REACTOME_VOLTAGE_GATED_POTASSIUM_CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 2.3 | REACTOME_DARPP_32_EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 1.4 | REACTOME_POTASSIUM_CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 1.3 | REACTOME_CELL_EXTRACELLULAR_MATRIX_INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.9 | REACTOME_RETROGRADE_NEUROTROPHIN_SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.6 | REACTOME_ACTIVATION_OF_THE_AP1_FAMILY_OF_TRANSCRIPTION_FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.4 | REACTOME_TRAFFICKING_AND_PROCESSING_OF_ENDOSOMAL_TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.4 | REACTOME_IKK_COMPLEX_RECRUITMENT_MEDIATED_BY_RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.9 | REACTOME_METABOLISM_OF_PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.9 | REACTOME_CASPASE_MEDIATED_CLEAVAGE_OF_CYTOSKELETAL_PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.9 | REACTOME_KERATAN_SULFATE_BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 1.3 | REACTOME_G_PROTEIN_ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 0.3 | REACTOME_EXTRINSIC_PATHWAY_FOR_APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.9 | REACTOME_RNA_POL_III_TRANSCRIPTION_TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.5 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_GOLGI_MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.9 | REACTOME_INTRINSIC_PATHWAY_FOR_APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.6 | REACTOME_PHASE1_FUNCTIONALIZATION_OF_COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 0.4 | REACTOME_IMMUNOREGULATORY_INTERACTIONS_BETWEEN_A_LYMPHOID_AND_A_NON_LYMPHOID_CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 1.1 | REACTOME_GOLGI_ASSOCIATED_VESICLE_BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.2 | REACTOME_FACILITATIVE_NA_INDEPENDENT_GLUCOSE_TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.2 | REACTOME_PURINE_RIBONUCLEOSIDE_MONOPHOSPHATE_BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 2.9 | REACTOME_SIGNALING_BY_RHO_GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.3 | REACTOME_REGULATION_OF_SIGNALING_BY_CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.5 | REACTOME_ION_TRANSPORT_BY_P_TYPE_ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 1.5 | REACTOME_TRANSPORT_OF_INORGANIC_CATIONS_ANIONS_AND_AMINO_ACIDS_OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.1 | REACTOME_PASSIVE_TRANSPORT_BY_AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.2 | REACTOME_ABCA_TRANSPORTERS_IN_LIPID_HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.1 | REACTOME_ABACAVIR_TRANSPORT_AND_METABOLISM | Genes involved in Abacavir transport and metabolism |