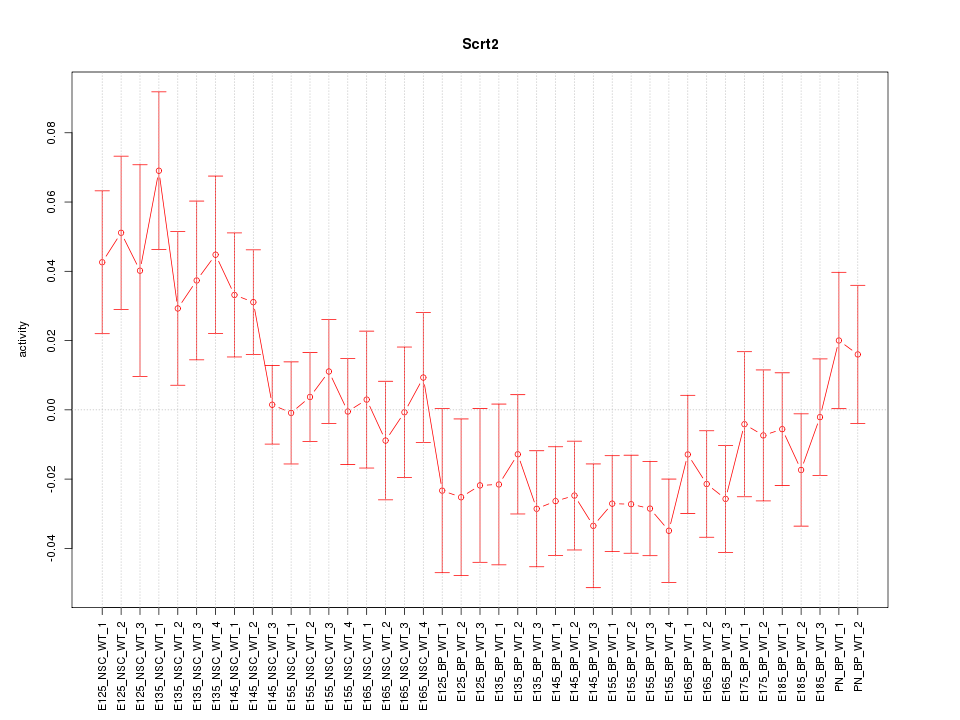
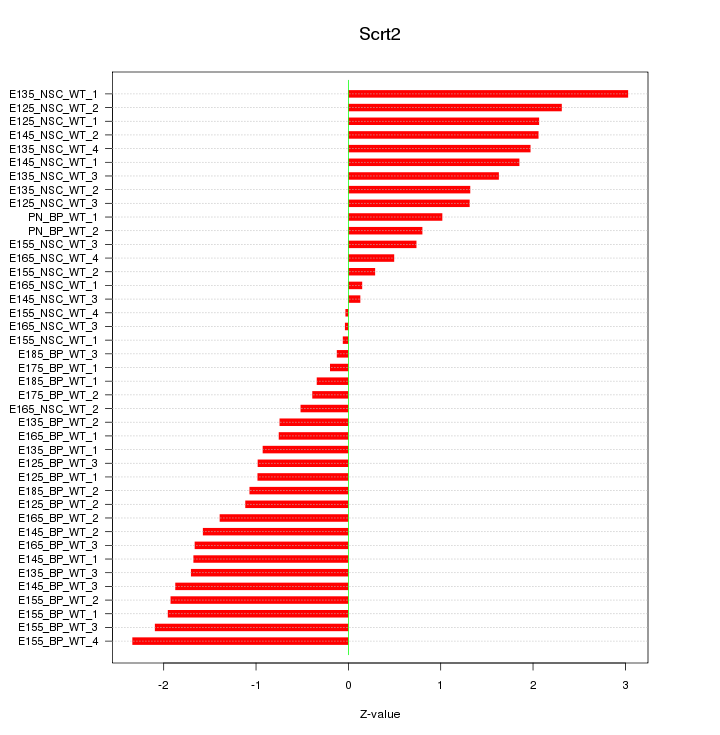
Motif ID: Scrt2
Z-value: 1.395
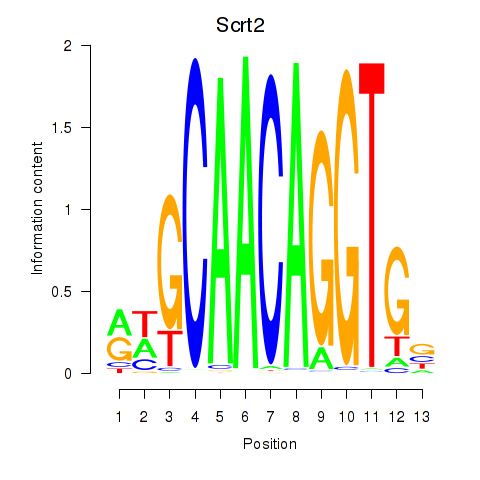
Transcription factors associated with Scrt2:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Scrt2 | ENSMUSG00000060257.2 | Scrt2 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Scrt2 | mm10_v2_chr2_+_152081529_152081624 | -0.74 | 2.6e-08 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.9 | 29.7 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 3.9 | 11.6 | GO:0003104 | positive regulation of glomerular filtration(GO:0003104) cell communication by chemical coupling(GO:0010643) |
| 3.4 | 10.1 | GO:0072034 | renal vesicle induction(GO:0072034) |
| 2.5 | 12.6 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 2.3 | 16.3 | GO:0071699 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 2.0 | 10.0 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 1.6 | 4.8 | GO:2000564 | CD8-positive, alpha-beta T cell proliferation(GO:0035740) negative regulation of regulatory T cell differentiation(GO:0045590) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) |
| 1.3 | 3.9 | GO:0002159 | desmosome assembly(GO:0002159) adherens junction maintenance(GO:0034334) intermediate filament bundle assembly(GO:0045110) maintenance of organ identity(GO:0048496) |
| 1.3 | 3.8 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 1.2 | 3.7 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 1.1 | 11.6 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.9 | 10.4 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.9 | 3.5 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.8 | 2.3 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.7 | 5.2 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.7 | 3.6 | GO:0032901 | positive regulation of neurotrophin production(GO:0032901) |
| 0.6 | 3.1 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.6 | 14.5 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.6 | 7.3 | GO:0046643 | regulation of gamma-delta T cell differentiation(GO:0045586) regulation of gamma-delta T cell activation(GO:0046643) |
| 0.4 | 3.9 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.4 | 2.6 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.4 | 2.0 | GO:0015822 | ornithine transport(GO:0015822) |
| 0.4 | 3.7 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.3 | 11.2 | GO:0001937 | negative regulation of endothelial cell proliferation(GO:0001937) |
| 0.3 | 1.8 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.3 | 5.0 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.3 | 3.6 | GO:0045187 | regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.2 | 5.5 | GO:0034142 | toll-like receptor 4 signaling pathway(GO:0034142) |
| 0.2 | 3.9 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.2 | 4.8 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.2 | 5.5 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.2 | 1.1 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.2 | 4.2 | GO:0008105 | asymmetric protein localization(GO:0008105) |
| 0.2 | 6.5 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.2 | 1.9 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.2 | 5.0 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.2 | 1.3 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.2 | 1.0 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.1 | 1.8 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 | 1.8 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.1 | 4.4 | GO:0042168 | heme metabolic process(GO:0042168) |
| 0.1 | 1.9 | GO:0046599 | regulation of centriole replication(GO:0046599) |
| 0.1 | 3.0 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.1 | 0.3 | GO:1901252 | regulation of intracellular transport of viral material(GO:1901252) |
| 0.1 | 0.3 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.1 | 0.3 | GO:0008655 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.1 | 0.2 | GO:0035696 | monocyte extravasation(GO:0035696) regulation of monocyte extravasation(GO:2000437) |
| 0.1 | 3.0 | GO:0031102 | neuron projection regeneration(GO:0031102) response to axon injury(GO:0048678) |
| 0.1 | 0.7 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 1.4 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 1.1 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.0 | 0.3 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.3 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.7 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.3 | GO:0045742 | positive regulation of epidermal growth factor receptor signaling pathway(GO:0045742) |
| 0.0 | 0.8 | GO:0051353 | positive regulation of oxidoreductase activity(GO:0051353) |
| 0.0 | 2.8 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.0 | 29.7 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 2.8 | 11.2 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 1.1 | 11.6 | GO:0005916 | fascia adherens(GO:0005916) |
| 1.0 | 11.6 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.9 | 5.5 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.8 | 3.9 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.4 | 3.1 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.4 | 1.9 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.4 | 3.9 | GO:0030057 | desmosome(GO:0030057) |
| 0.3 | 13.7 | GO:0043034 | costamere(GO:0043034) |
| 0.2 | 3.7 | GO:0042555 | MCM complex(GO:0042555) |
| 0.2 | 1.8 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.2 | 5.0 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 3.1 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 23.6 | GO:0044798 | nuclear transcription factor complex(GO:0044798) |
| 0.1 | 1.3 | GO:0000243 | commitment complex(GO:0000243) |
| 0.1 | 1.1 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 2.3 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 16.1 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.1 | 0.3 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.1 | 2.6 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 5.5 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 11.6 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 4.4 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 6.3 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 1.1 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 1.8 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 8.4 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.4 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 4.4 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.8 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.7 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 11.6 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) gap junction channel activity involved in cell communication by electrical coupling(GO:1903763) |
| 2.3 | 11.6 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 2.1 | 29.7 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 2.0 | 10.0 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 1.3 | 5.0 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 1.1 | 14.5 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.8 | 16.1 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.6 | 1.8 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.5 | 2.0 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.5 | 1.4 | GO:0019002 | GMP binding(GO:0019002) |
| 0.5 | 3.6 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.4 | 5.5 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.3 | 5.5 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.3 | 3.9 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.3 | 3.8 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.3 | 1.8 | GO:0071253 | connexin binding(GO:0071253) |
| 0.2 | 3.7 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.2 | 11.2 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.2 | 0.8 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.2 | 4.5 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.2 | 3.1 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.1 | 10.4 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.1 | 3.1 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 1.8 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 9.4 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 10.1 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.1 | 3.7 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.1 | 2.3 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 5.3 | GO:0020037 | heme binding(GO:0020037) |
| 0.1 | 1.1 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.1 | 0.3 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 12.6 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.1 | 4.8 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 9.7 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 2.6 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 2.2 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 3.7 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.3 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 1.3 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.7 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.3 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.7 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 13.4 | GO:0000977 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) |
| 0.0 | 0.3 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 6.0 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.6 | GO:0016504 | peptidase activator activity(GO:0016504) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 14.2 | NABA_COLLAGENS | Genes encoding collagen proteins |
| 0.3 | 10.0 | PID_SYNDECAN_4_PATHWAY | Syndecan-4-mediated signaling events |
| 0.3 | 11.6 | PID_NCADHERIN_PATHWAY | N-cadherin signaling events |
| 0.3 | 3.9 | PID_INTEGRIN2_PATHWAY | Beta2 integrin cell surface interactions |
| 0.2 | 10.4 | PID_FANCONI_PATHWAY | Fanconi anemia pathway |
| 0.2 | 4.8 | PID_IL12_STAT4_PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 3.7 | PID_ATR_PATHWAY | ATR signaling pathway |
| 0.1 | 11.0 | NABA_ECM_GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 3.4 | PID_RAC1_REG_PATHWAY | Regulation of RAC1 activity |
| 0.0 | 1.8 | PID_ECADHERIN_NASCENT_AJ_PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 2.3 | PID_ERA_GENOMIC_PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 2.2 | PID_MTOR_4PATHWAY | mTOR signaling pathway |
| 0.0 | 3.7 | NABA_SECRETED_FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.4 | NABA_ECM_AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 10.4 | REACTOME_REGULATION_OF_THE_FANCONI_ANEMIA_PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 1.0 | 11.6 | REACTOME_GAP_JUNCTION_ASSEMBLY | Genes involved in Gap junction assembly |
| 0.4 | 10.0 | REACTOME_HS_GAG_DEGRADATION | Genes involved in HS-GAG degradation |
| 0.3 | 3.5 | REACTOME_VEGF_LIGAND_RECEPTOR_INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.3 | 1.8 | REACTOME_APOPTOTIC_CLEAVAGE_OF_CELL_ADHESION_PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.2 | 14.8 | REACTOME_COLLAGEN_FORMATION | Genes involved in Collagen formation |
| 0.2 | 4.8 | REACTOME_TRAF6_MEDIATED_IRF7_ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.2 | 3.7 | REACTOME_UNWINDING_OF_DNA | Genes involved in Unwinding of DNA |
| 0.1 | 3.8 | REACTOME_STRIATED_MUSCLE_CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 5.0 | REACTOME_PACKAGING_OF_TELOMERE_ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.1 | 3.1 | REACTOME_DEADENYLATION_OF_MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 9.2 | REACTOME_RESPONSE_TO_ELEVATED_PLATELET_CYTOSOLIC_CA2_ | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 3.1 | REACTOME_TIGHT_JUNCTION_INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 2.0 | REACTOME_AMINO_ACID_TRANSPORT_ACROSS_THE_PLASMA_MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 3.9 | REACTOME_INTEGRIN_CELL_SURFACE_INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 3.4 | REACTOME_FACTORS_INVOLVED_IN_MEGAKARYOCYTE_DEVELOPMENT_AND_PLATELET_PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.3 | REACTOME_PYRIMIDINE_METABOLISM | Genes involved in Pyrimidine metabolism |