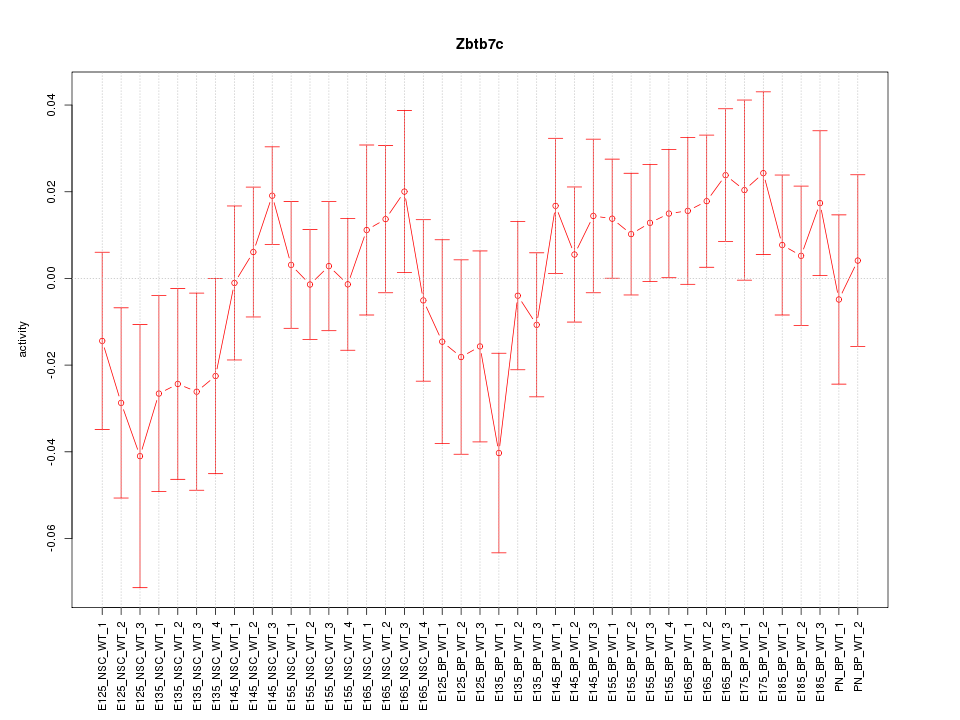
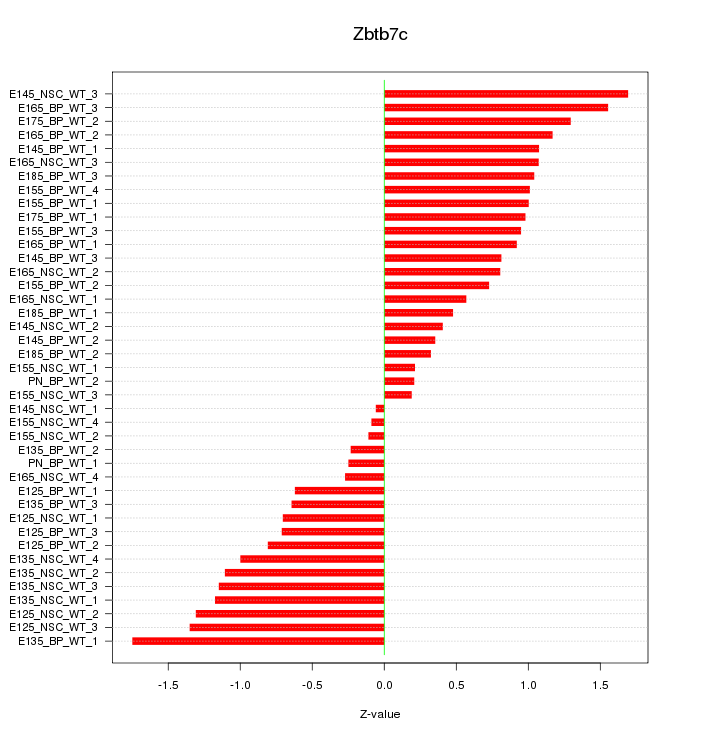
Motif ID: Zbtb7c
Z-value: 0.905
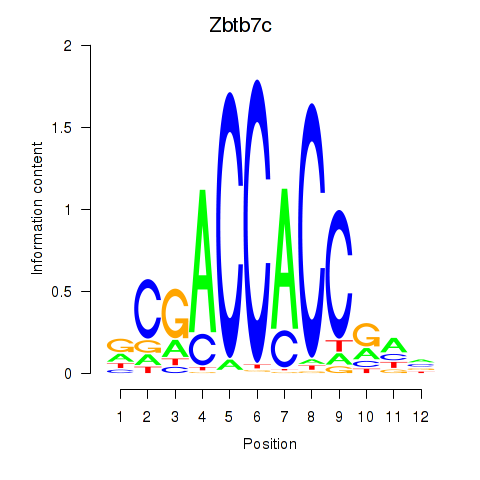
Transcription factors associated with Zbtb7c:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Zbtb7c | ENSMUSG00000044646.8 | Zbtb7c |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zbtb7c | mm10_v2_chr18_+_76059458_76059501 | 0.12 | 4.4e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.1 | GO:1902022 | L-lysine transport(GO:1902022) |
| 1.3 | 11.4 | GO:1900034 | regulation of cellular response to heat(GO:1900034) |
| 0.7 | 2.1 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.6 | 1.9 | GO:0098928 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 0.6 | 3.9 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) regulation of mitophagy in response to mitochondrial depolarization(GO:1904923) |
| 0.6 | 4.4 | GO:0099640 | axo-dendritic protein transport(GO:0099640) |
| 0.6 | 1.7 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.5 | 2.1 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.4 | 1.4 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.4 | 4.7 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.4 | 5.7 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.3 | 1.0 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.3 | 0.9 | GO:0061181 | regulation of chondrocyte development(GO:0061181) |
| 0.3 | 1.5 | GO:0035864 | response to potassium ion(GO:0035864) cellular response to potassium ion(GO:0035865) |
| 0.3 | 0.9 | GO:0007521 | muscle cell fate determination(GO:0007521) mammary placode formation(GO:0060596) |
| 0.3 | 2.1 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.3 | 1.6 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.3 | 2.1 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.2 | 6.5 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.2 | 2.2 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.2 | 1.2 | GO:1903056 | positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.2 | 1.8 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) |
| 0.2 | 1.3 | GO:0030242 | pexophagy(GO:0030242) |
| 0.2 | 0.5 | GO:0015747 | urate transport(GO:0015747) |
| 0.2 | 1.8 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.2 | 0.8 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.2 | 1.6 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.2 | 0.9 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.1 | 3.2 | GO:0014898 | muscle hypertrophy in response to stress(GO:0003299) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.1 | 8.5 | GO:0015807 | L-amino acid transport(GO:0015807) |
| 0.1 | 0.9 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.1 | 1.6 | GO:0035646 | endosome to melanosome transport(GO:0035646) endosome to pigment granule transport(GO:0043485) pigment granule maturation(GO:0048757) |
| 0.1 | 0.8 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.1 | 2.5 | GO:0021702 | cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.1 | 1.7 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.1 | 0.9 | GO:2000465 | regulation of glycogen (starch) synthase activity(GO:2000465) |
| 0.1 | 0.5 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.1 | 2.3 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.1 | 0.9 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.1 | 0.2 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.2 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.0 | 0.6 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.5 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 3.0 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 1.1 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 1.2 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 | 0.3 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 6.3 | GO:0071805 | potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 0.2 | GO:2001170 | negative regulation of ATP biosynthetic process(GO:2001170) |
| 0.0 | 0.9 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.3 | GO:0044539 | long-chain fatty acid import(GO:0044539) |
| 0.0 | 0.6 | GO:0045742 | positive regulation of epidermal growth factor receptor signaling pathway(GO:0045742) positive regulation of ERBB signaling pathway(GO:1901186) |
| 0.0 | 0.1 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.0 | 1.7 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.0 | 0.4 | GO:0010831 | positive regulation of myotube differentiation(GO:0010831) |
| 0.0 | 6.6 | GO:0007409 | axonogenesis(GO:0007409) |
| 0.0 | 3.9 | GO:0007059 | chromosome segregation(GO:0007059) |
| 0.0 | 0.2 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.4 | GO:0001706 | endoderm formation(GO:0001706) negative regulation of DNA biosynthetic process(GO:2000279) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 11.4 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.9 | 3.5 | GO:0043259 | laminin-1 complex(GO:0005606) laminin-10 complex(GO:0043259) |
| 0.6 | 1.9 | GO:1990257 | piccolo-bassoon transport vesicle(GO:1990257) |
| 0.4 | 2.3 | GO:0008091 | spectrin(GO:0008091) |
| 0.4 | 3.7 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.4 | 2.9 | GO:0000836 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.4 | 1.8 | GO:0005587 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.2 | 4.4 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.2 | 1.8 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.2 | 6.5 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.2 | 1.7 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.2 | 0.6 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.1 | 4.6 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 2.1 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 1.2 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 0.8 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.1 | 1.3 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 1.8 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.1 | 0.3 | GO:0097651 | phosphatidylinositol 3-kinase complex, class I(GO:0097651) |
| 0.1 | 6.1 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 2.2 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 1.6 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 5.7 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.2 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.6 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 2.9 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.3 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.2 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 3.2 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.9 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 4.7 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 8.7 | GO:0016604 | nuclear body(GO:0016604) |
| 0.0 | 0.5 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 1.4 | GO:0036064 | ciliary basal body(GO:0036064) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 11.4 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 1.0 | 4.1 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.5 | 5.7 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.4 | 3.5 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.4 | 1.2 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.3 | 1.0 | GO:0008732 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.3 | 2.2 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.2 | 0.9 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.2 | 1.6 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.2 | 2.8 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.2 | 2.7 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.2 | 8.5 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.2 | 2.9 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.1 | 1.9 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.1 | 4.7 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 2.1 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.1 | 0.5 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 0.4 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 3.5 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 3.5 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 0.6 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.1 | 0.3 | GO:0043758 | acetate-CoA ligase (ADP-forming) activity(GO:0043758) |
| 0.1 | 2.1 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 1.2 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.1 | 2.3 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 1.1 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.1 | 1.6 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.1 | 0.5 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) |
| 0.1 | 1.7 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.1 | 1.2 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.9 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.5 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.0 | 0.6 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.6 | GO:0030553 | cGMP binding(GO:0030553) |
| 0.0 | 0.3 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 2.3 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 4.2 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.3 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 4.0 | GO:0016810 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds(GO:0016810) |
| 0.0 | 1.7 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.2 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.2 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.0 | 4.4 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.3 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 1.8 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 1.3 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 0.9 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.2 | GO:0005521 | lamin binding(GO:0005521) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 11.4 | PID_P38_GAMMA_DELTA_PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.4 | 3.5 | PID_INTEGRIN4_PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.2 | 6.4 | NABA_BASEMENT_MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 5.7 | PID_PI3K_PLC_TRK_PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.6 | PID_CONE_PATHWAY | Visual signal transduction: Cones |
| 0.0 | 1.6 | PID_ECADHERIN_NASCENT_AJ_PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.9 | PID_BETA_CATENIN_DEG_PATHWAY | Degradation of beta catenin |
| 0.0 | 2.1 | PID_ARF6_TRAFFICKING_PATHWAY | Arf6 trafficking events |
| 0.0 | 2.1 | PID_AR_PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 1.9 | NABA_ECM_GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.3 | PID_TELOMERASE_PATHWAY | Regulation of Telomerase |
| 0.0 | 0.4 | ST_P38_MAPK_PATHWAY | p38 MAPK Pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 11.4 | REACTOME_CASPASE_MEDIATED_CLEAVAGE_OF_CYTOSKELETAL_PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.3 | 12.6 | REACTOME_AMINO_ACID_TRANSPORT_ACROSS_THE_PLASMA_MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.2 | 3.5 | REACTOME_NEF_MEDIATED_DOWNREGULATION_OF_MHC_CLASS_I_COMPLEX_CELL_SURFACE_EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.2 | 4.0 | REACTOME_CRMPS_IN_SEMA3A_SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.2 | 4.4 | REACTOME_KINESINS | Genes involved in Kinesins |
| 0.1 | 5.7 | REACTOME_CA_DEPENDENT_EVENTS | Genes involved in Ca-dependent events |
| 0.1 | 5.1 | REACTOME_VOLTAGE_GATED_POTASSIUM_CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 1.6 | REACTOME_SYNTHESIS_OF_SUBSTRATES_IN_N_GLYCAN_BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 0.5 | REACTOME_ORGANIC_CATION_ANION_ZWITTERION_TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 2.3 | REACTOME_INTERACTION_BETWEEN_L1_AND_ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 5.3 | REACTOME_INTEGRIN_CELL_SURFACE_INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 3.1 | REACTOME_MEIOTIC_SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 2.2 | REACTOME_TRANSPORT_OF_INORGANIC_CATIONS_ANIONS_AND_AMINO_ACIDS_OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.8 | REACTOME_APOPTOTIC_CLEAVAGE_OF_CELLULAR_PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.3 | REACTOME_ALPHA_LINOLENIC_ACID_ALA_METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.3 | REACTOME_KERATAN_SULFATE_BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |