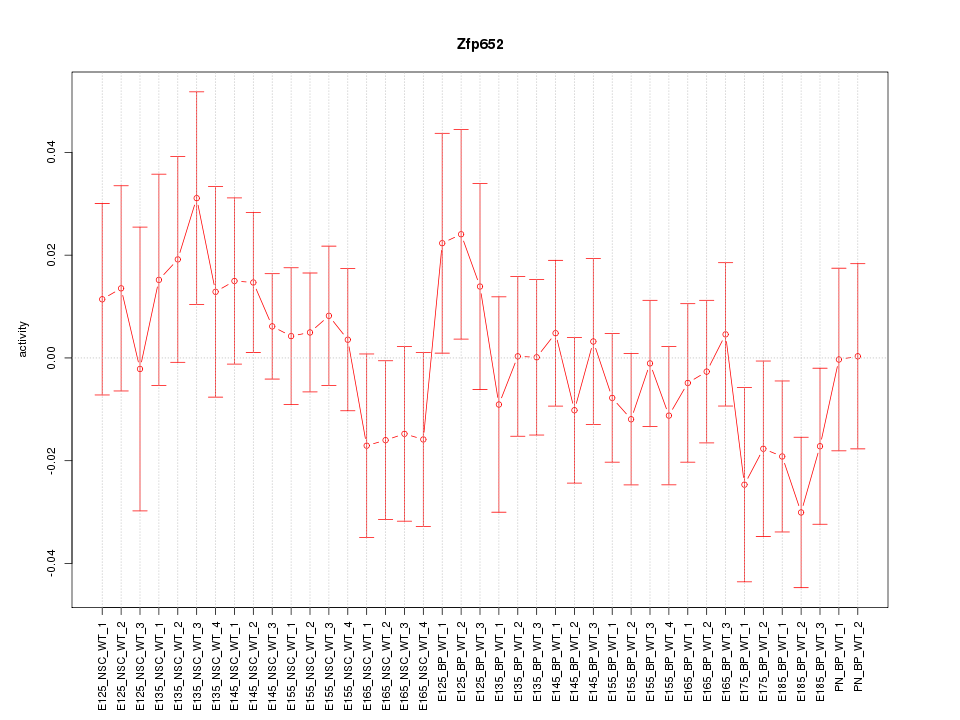
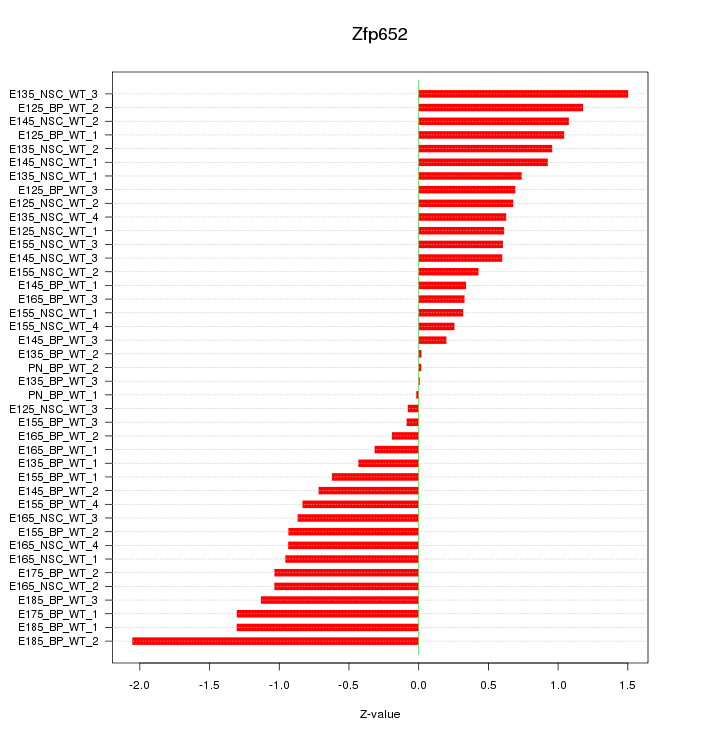
Motif ID: Zfp652
Z-value: 0.821
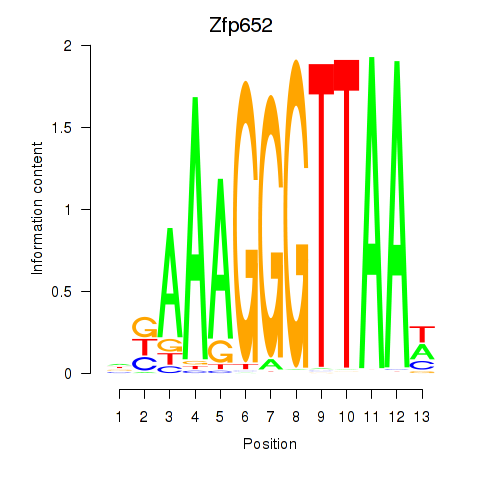
Transcription factors associated with Zfp652:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Zfp652 | ENSMUSG00000075595.3 | Zfp652 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zfp652 | mm10_v2_chr11_+_95712673_95712673 | -0.23 | 1.5e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 10.0 | GO:0090274 | positive regulation of somatostatin secretion(GO:0090274) |
| 2.2 | 6.6 | GO:0021557 | oculomotor nerve development(GO:0021557) |
| 1.1 | 15.6 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 1.0 | 3.1 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 1.0 | 3.0 | GO:1904020 | regulation of G-protein coupled receptor internalization(GO:1904020) |
| 1.0 | 2.9 | GO:0097350 | neutrophil clearance(GO:0097350) |
| 0.7 | 3.6 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.6 | 3.2 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.5 | 2.1 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.5 | 1.5 | GO:0009174 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.4 | 2.2 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.4 | 1.7 | GO:0060032 | notochord regression(GO:0060032) |
| 0.4 | 1.5 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.4 | 1.5 | GO:0021502 | neural fold elevation formation(GO:0021502) |
| 0.4 | 1.1 | GO:0086053 | AV node cell to bundle of His cell communication by electrical coupling(GO:0086053) |
| 0.3 | 1.0 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 0.2 | 2.3 | GO:0061470 | interleukin-21 production(GO:0032625) T follicular helper cell differentiation(GO:0061470) interleukin-21 secretion(GO:0072619) |
| 0.2 | 1.8 | GO:0015074 | DNA integration(GO:0015074) |
| 0.2 | 3.6 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.2 | 1.8 | GO:0061032 | visceral serous pericardium development(GO:0061032) |
| 0.2 | 0.9 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.2 | 0.5 | GO:0061181 | regulation of chondrocyte development(GO:0061181) |
| 0.1 | 1.5 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.1 | 1.6 | GO:0045618 | positive regulation of keratinocyte differentiation(GO:0045618) |
| 0.1 | 1.5 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 1.1 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.1 | 2.1 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.1 | 0.2 | GO:0035672 | transepithelial chloride transport(GO:0030321) oligopeptide transmembrane transport(GO:0035672) |
| 0.1 | 0.3 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.1 | 1.6 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.1 | 0.4 | GO:2001054 | negative regulation of mesenchymal cell apoptotic process(GO:2001054) |
| 0.1 | 0.6 | GO:0019065 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.1 | 0.4 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.1 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 1.6 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.0 | 0.1 | GO:1903224 | regulation of endodermal cell differentiation(GO:1903224) |
| 0.0 | 0.1 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 2.1 | GO:0043392 | negative regulation of DNA binding(GO:0043392) |
| 0.0 | 0.7 | GO:0035115 | embryonic forelimb morphogenesis(GO:0035115) |
| 0.0 | 6.4 | GO:0043524 | negative regulation of neuron apoptotic process(GO:0043524) |
| 0.0 | 0.1 | GO:0002667 | lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) |
| 0.0 | 0.9 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 1.0 | GO:0070306 | lens fiber cell differentiation(GO:0070306) |
| 0.0 | 0.3 | GO:0022038 | corpus callosum development(GO:0022038) |
| 0.0 | 0.6 | GO:0003170 | heart valve development(GO:0003170) |
| 0.0 | 0.1 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.1 | GO:0032782 | bile acid secretion(GO:0032782) |
| 0.0 | 0.4 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.2 | GO:0014823 | response to activity(GO:0014823) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 11.6 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.2 | 1.0 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.2 | 0.9 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.1 | 1.7 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.1 | 8.0 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 2.9 | GO:0043657 | host(GO:0018995) host cell part(GO:0033643) host cell(GO:0043657) |
| 0.1 | 5.0 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 1.6 | GO:0031105 | septin complex(GO:0031105) |
| 0.1 | 1.1 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 1.4 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 10.0 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 0.6 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.1 | 3.4 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 0.9 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 0.4 | GO:0071664 | beta-catenin-TCF7L2 complex(GO:0070369) catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 2.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 3.4 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.2 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 3.7 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 1.6 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.1 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 1.6 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.9 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.6 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.2 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.3 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 2.1 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 1.0 | 9.0 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.5 | 1.6 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.5 | 1.8 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.4 | 15.6 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.4 | 9.7 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.4 | 1.1 | GO:0086077 | gap junction channel activity involved in AV node cell-bundle of His cell electrical coupling(GO:0086077) |
| 0.4 | 2.1 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.3 | 1.5 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.3 | 2.9 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.3 | 3.1 | GO:0035014 | retinoic acid binding(GO:0001972) phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.2 | 1.5 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.2 | 1.7 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.2 | 3.0 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.2 | 10.0 | GO:0008528 | G-protein coupled peptide receptor activity(GO:0008528) |
| 0.2 | 0.9 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.2 | 2.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.2 | 2.1 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.1 | 1.5 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.1 | 1.5 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 1.1 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 0.2 | GO:0015563 | uptake transmembrane transporter activity(GO:0015563) |
| 0.0 | 2.2 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 1.7 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.3 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 1.1 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.4 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.0 | 3.2 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.4 | GO:0070016 | gamma-catenin binding(GO:0045295) armadillo repeat domain binding(GO:0070016) |
| 0.0 | 2.3 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 1.0 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.2 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.9 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.4 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.5 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.2 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.6 | GO:0031593 | polyubiquitin binding(GO:0031593) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 9.5 | PID_DELTA_NP63_PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 1.8 | PID_INTEGRIN5_PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 3.1 | PID_RXR_VDR_PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 3.1 | SA_PTEN_PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.1 | 2.2 | PID_ARF_3PATHWAY | Arf1 pathway |
| 0.1 | 3.2 | PID_P38_ALPHA_BETA_PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.1 | 3.4 | PID_RHOA_PATHWAY | RhoA signaling pathway |
| 0.1 | 1.7 | PID_HEDGEHOG_2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.5 | ST_INTERFERON_GAMMA_PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.9 | PID_ATM_PATHWAY | ATM pathway |
| 0.0 | 1.0 | PID_NFAT_TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.5 | PID_BETA_CATENIN_DEG_PATHWAY | Degradation of beta catenin |
| 0.0 | 0.6 | PID_ERBB1_INTERNALIZATION_PATHWAY | Internalization of ErbB1 |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.1 | REACTOME_P2Y_RECEPTORS | Genes involved in P2Y receptors |
| 0.2 | 15.6 | REACTOME_G_ALPHA_I_SIGNALLING_EVENTS | Genes involved in G alpha (i) signalling events |
| 0.2 | 5.5 | REACTOME_REGULATION_OF_BETA_CELL_DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.1 | 10.0 | REACTOME_PEPTIDE_LIGAND_BINDING_RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 1.1 | REACTOME_GAP_JUNCTION_ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 1.8 | REACTOME_IMMUNOREGULATORY_INTERACTIONS_BETWEEN_A_LYMPHOID_AND_A_NON_LYMPHOID_CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 2.9 | REACTOME_VOLTAGE_GATED_POTASSIUM_CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 1.5 | REACTOME_PYRIMIDINE_METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.1 | 1.5 | REACTOME_GLUTATHIONE_CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 1.6 | REACTOME_PRE_NOTCH_TRANSCRIPTION_AND_TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.1 | 0.9 | REACTOME_HOMOLOGOUS_RECOMBINATION_REPAIR_OF_REPLICATION_INDEPENDENT_DOUBLE_STRAND_BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 3.1 | REACTOME_NUCLEAR_RECEPTOR_TRANSCRIPTION_PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.5 | REACTOME_REGULATION_OF_IFNG_SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 1.0 | REACTOME_STRIATED_MUSCLE_CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.1 | REACTOME_CD28_DEPENDENT_VAV1_PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.6 | REACTOME_EGFR_DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.1 | REACTOME_PASSIVE_TRANSPORT_BY_AQUAPORINS | Genes involved in Passive Transport by Aquaporins |