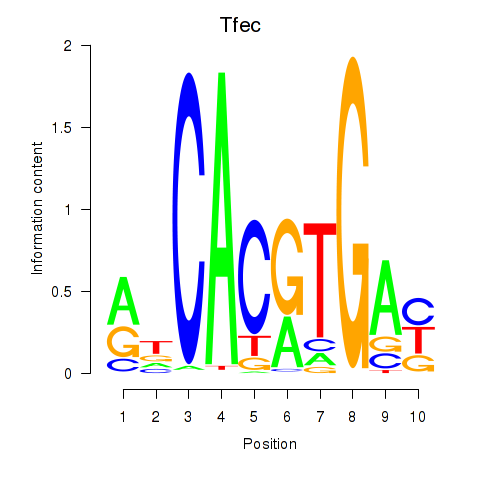
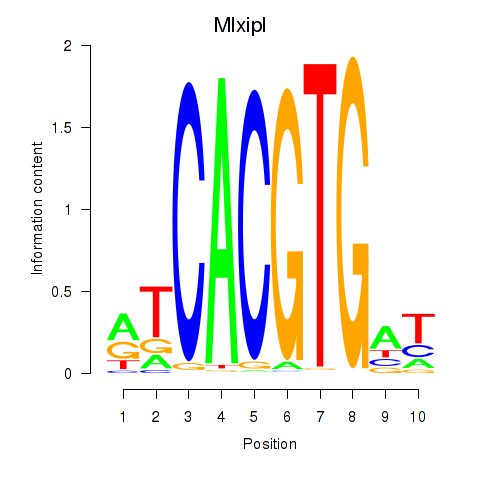
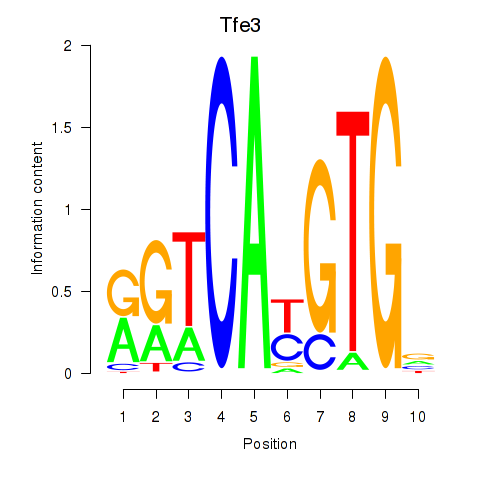
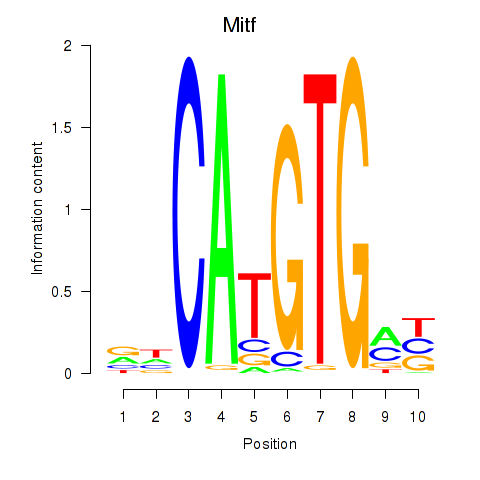
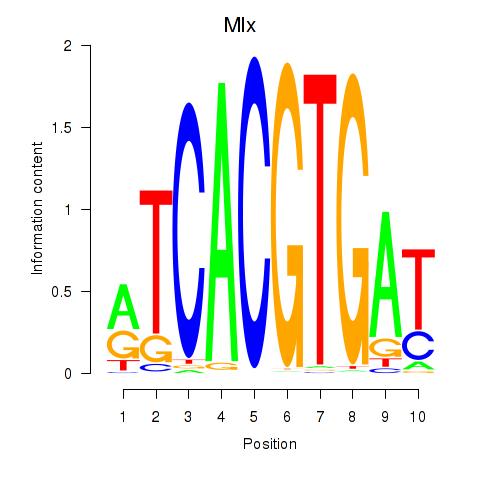
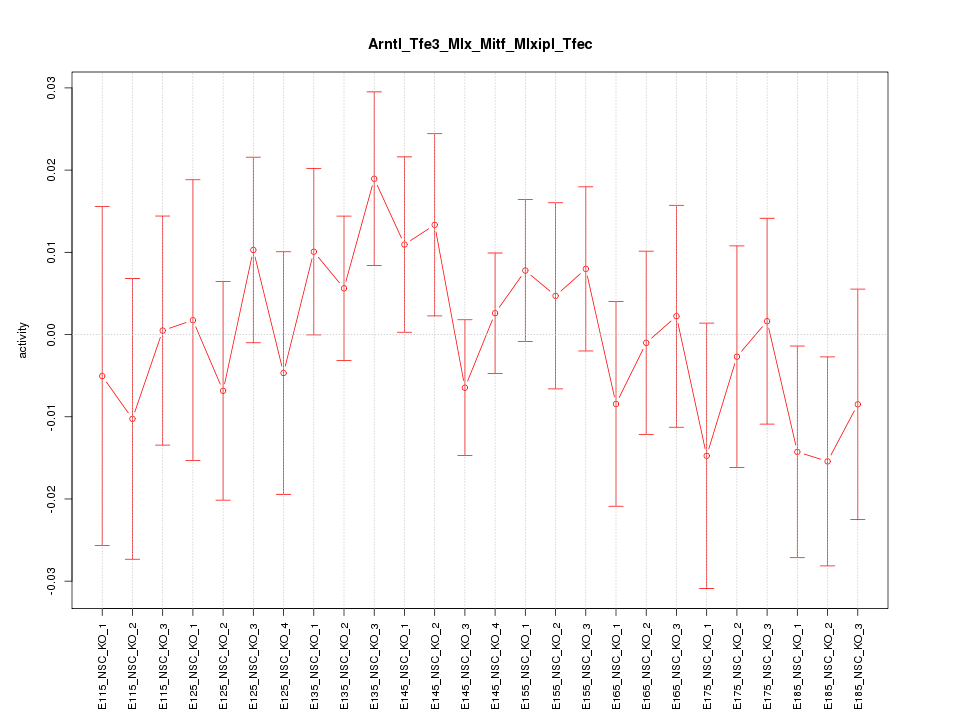
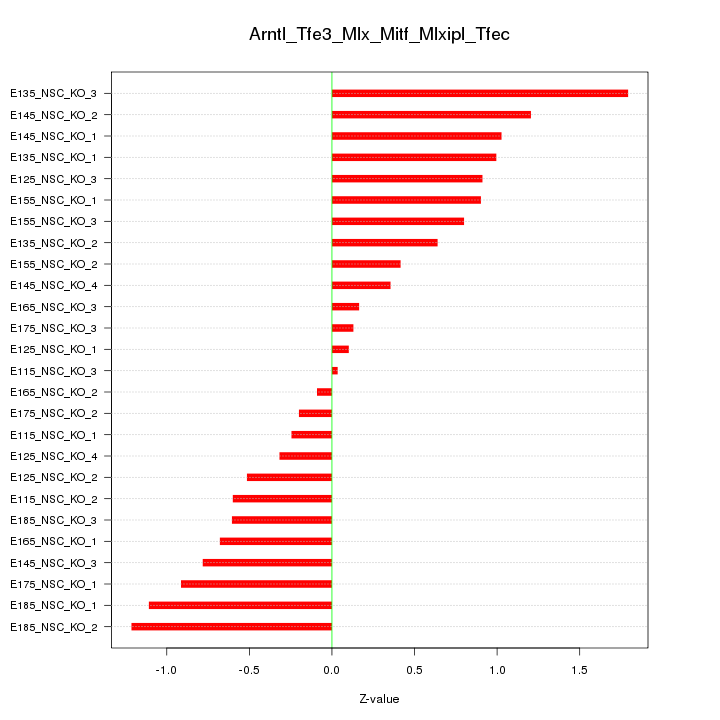
| 1.2 |
4.7 |
GO:0010288 |
response to lead ion(GO:0010288) |
| 0.6 |
4.9 |
GO:0071557 |
histone H3-K27 demethylation(GO:0071557) |
| 0.6 |
1.7 |
GO:0007621 |
negative regulation of female receptivity(GO:0007621) |
| 0.5 |
0.5 |
GO:0061470 |
T follicular helper cell differentiation(GO:0061470) |
| 0.5 |
2.3 |
GO:0014022 |
neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.5 |
1.4 |
GO:0010845 |
positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.4 |
1.3 |
GO:0032053 |
ciliary basal body organization(GO:0032053) |
| 0.3 |
1.3 |
GO:0034184 |
positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.3 |
1.2 |
GO:0070829 |
heterochromatin maintenance(GO:0070829) |
| 0.3 |
1.8 |
GO:0007253 |
cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.3 |
0.9 |
GO:0072070 |
establishment or maintenance of polarity of embryonic epithelium(GO:0016332) loop of Henle development(GO:0072070) metanephric loop of Henle development(GO:0072236) |
| 0.3 |
2.7 |
GO:1902231 |
positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.3 |
0.3 |
GO:0070827 |
chromatin maintenance(GO:0070827) |
| 0.3 |
0.8 |
GO:0036091 |
positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.3 |
0.8 |
GO:0072429 |
response to cell cycle checkpoint signaling(GO:0072396) response to DNA integrity checkpoint signaling(GO:0072402) response to DNA damage checkpoint signaling(GO:0072423) response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.3 |
2.5 |
GO:0048742 |
regulation of skeletal muscle fiber development(GO:0048742) |
| 0.2 |
1.5 |
GO:0046502 |
uroporphyrinogen III metabolic process(GO:0046502) |
| 0.2 |
0.7 |
GO:0006285 |
base-excision repair, AP site formation(GO:0006285) |
| 0.2 |
1.4 |
GO:1904715 |
negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 0.2 |
1.1 |
GO:0090273 |
regulation of somatostatin secretion(GO:0090273) |
| 0.2 |
1.3 |
GO:0006570 |
tyrosine metabolic process(GO:0006570) |
| 0.2 |
0.6 |
GO:0046032 |
ADP catabolic process(GO:0046032) |
| 0.2 |
0.6 |
GO:0010248 |
B cell negative selection(GO:0002352) establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.2 |
1.3 |
GO:0043137 |
DNA replication, removal of RNA primer(GO:0043137) |
| 0.2 |
0.6 |
GO:1904274 |
tricellular tight junction assembly(GO:1904274) |
| 0.2 |
0.8 |
GO:0042364 |
water-soluble vitamin biosynthetic process(GO:0042364) |
| 0.2 |
0.8 |
GO:0035752 |
lysosomal lumen pH elevation(GO:0035752) |
| 0.2 |
0.5 |
GO:2000620 |
positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.2 |
0.7 |
GO:0006545 |
glycine biosynthetic process(GO:0006545) |
| 0.2 |
0.7 |
GO:0030576 |
Cajal body organization(GO:0030576) |
| 0.2 |
0.5 |
GO:0030421 |
defecation(GO:0030421) |
| 0.2 |
0.8 |
GO:1900085 |
negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) |
| 0.2 |
0.8 |
GO:0035984 |
response to trichostatin A(GO:0035983) cellular response to trichostatin A(GO:0035984) |
| 0.2 |
0.6 |
GO:0035795 |
negative regulation of mitochondrial membrane permeability(GO:0035795) |
| 0.2 |
0.6 |
GO:0060729 |
intestinal epithelial structure maintenance(GO:0060729) |
| 0.1 |
2.9 |
GO:0071392 |
cellular response to estradiol stimulus(GO:0071392) |
| 0.1 |
1.1 |
GO:0051001 |
negative regulation of nitric-oxide synthase activity(GO:0051001) |
| 0.1 |
0.6 |
GO:0060032 |
notochord regression(GO:0060032) |
| 0.1 |
0.4 |
GO:0006419 |
alanyl-tRNA aminoacylation(GO:0006419) |
| 0.1 |
0.7 |
GO:0040038 |
polar body extrusion after meiotic divisions(GO:0040038) |
| 0.1 |
0.4 |
GO:0045472 |
response to ether(GO:0045472) |
| 0.1 |
0.4 |
GO:0070476 |
rRNA (guanine-N7)-methylation(GO:0070476) |
| 0.1 |
0.8 |
GO:0006046 |
N-acetylglucosamine catabolic process(GO:0006046) |
| 0.1 |
0.4 |
GO:2000508 |
regulation of dendritic cell chemotaxis(GO:2000508) positive regulation of dendritic cell chemotaxis(GO:2000510) |
| 0.1 |
0.9 |
GO:0061484 |
hematopoietic stem cell homeostasis(GO:0061484) |
| 0.1 |
0.2 |
GO:0006867 |
asparagine transport(GO:0006867) glutamine transport(GO:0006868) |
| 0.1 |
0.4 |
GO:0051902 |
negative regulation of mitochondrial depolarization(GO:0051902) negative regulation of membrane depolarization(GO:1904180) |
| 0.1 |
0.5 |
GO:0070459 |
prolactin secretion(GO:0070459) |
| 0.1 |
0.2 |
GO:0036166 |
phenotypic switching(GO:0036166) |
| 0.1 |
0.4 |
GO:2000049 |
positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.1 |
0.4 |
GO:1902219 |
regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902218) negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.1 |
2.4 |
GO:0071378 |
growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.1 |
0.4 |
GO:0007181 |
transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.1 |
0.4 |
GO:0009446 |
putrescine biosynthetic process(GO:0009446) |
| 0.1 |
0.5 |
GO:2000795 |
negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.1 |
0.5 |
GO:1901029 |
negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.1 |
0.5 |
GO:0051151 |
negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.1 |
0.1 |
GO:1904339 |
negative regulation of dopaminergic neuron differentiation(GO:1904339) |
| 0.1 |
0.4 |
GO:0051660 |
cortical microtubule organization(GO:0043622) establishment of centrosome localization(GO:0051660) |
| 0.1 |
1.2 |
GO:0006995 |
cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 |
0.5 |
GO:1904683 |
regulation of metalloendopeptidase activity(GO:1904683) negative regulation of metalloendopeptidase activity(GO:1904684) negative regulation of metallopeptidase activity(GO:1905049) |
| 0.1 |
0.7 |
GO:2000781 |
positive regulation of double-strand break repair(GO:2000781) |
| 0.1 |
0.3 |
GO:0048597 |
post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.1 |
0.4 |
GO:0002949 |
tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.1 |
0.7 |
GO:0031119 |
tRNA pseudouridine synthesis(GO:0031119) |
| 0.1 |
0.3 |
GO:0002940 |
tRNA N2-guanine methylation(GO:0002940) |
| 0.1 |
0.3 |
GO:0061144 |
alveolar secondary septum development(GO:0061144) |
| 0.1 |
0.5 |
GO:0006982 |
response to lipid hydroperoxide(GO:0006982) |
| 0.1 |
0.3 |
GO:0035864 |
response to potassium ion(GO:0035864) cellular response to potassium ion(GO:0035865) |
| 0.1 |
0.6 |
GO:0045218 |
zonula adherens maintenance(GO:0045218) |
| 0.1 |
0.4 |
GO:0090238 |
positive regulation of arachidonic acid secretion(GO:0090238) |
| 0.1 |
0.4 |
GO:0003376 |
sphingosine-1-phosphate signaling pathway(GO:0003376) sphingolipid mediated signaling pathway(GO:0090520) |
| 0.1 |
2.7 |
GO:0034723 |
DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 |
0.3 |
GO:1903774 |
positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.1 |
0.3 |
GO:0001827 |
inner cell mass cell fate commitment(GO:0001827) |
| 0.1 |
0.6 |
GO:0015808 |
L-alanine transport(GO:0015808) |
| 0.1 |
1.1 |
GO:1990126 |
retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.1 |
0.4 |
GO:0051661 |
maintenance of centrosome location(GO:0051661) |
| 0.1 |
0.4 |
GO:0021764 |
amygdala development(GO:0021764) |
| 0.1 |
0.4 |
GO:0018002 |
N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.1 |
1.2 |
GO:1901741 |
positive regulation of myoblast fusion(GO:1901741) |
| 0.1 |
0.6 |
GO:0010944 |
negative regulation of transcription by competitive promoter binding(GO:0010944) |
| 0.1 |
0.5 |
GO:0035428 |
hexose transmembrane transport(GO:0035428) |
| 0.1 |
0.2 |
GO:0060672 |
epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.1 |
0.2 |
GO:0072139 |
glomerular parietal epithelial cell differentiation(GO:0072139) |
| 0.1 |
0.5 |
GO:0033169 |
histone H3-K9 demethylation(GO:0033169) |
| 0.1 |
0.2 |
GO:0072554 |
arterial endothelial cell fate commitment(GO:0060844) blood vessel endothelial cell fate commitment(GO:0060846) endothelial cell fate specification(GO:0060847) Notch signaling pathway involved in arterial endothelial cell fate commitment(GO:0060853) blood vessel lumenization(GO:0072554) blood vessel endothelial cell fate specification(GO:0097101) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) positive regulation of heart induction(GO:1901321) regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) positive regulation of cell proliferation involved in heart morphogenesis(GO:2000138) |
| 0.1 |
0.4 |
GO:1905098 |
negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.1 |
0.3 |
GO:1904995 |
negative regulation of leukocyte adhesion to vascular endothelial cell(GO:1904995) |
| 0.1 |
0.2 |
GO:0007354 |
zygotic determination of anterior/posterior axis, embryo(GO:0007354) |
| 0.1 |
0.2 |
GO:0036353 |
histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.1 |
0.2 |
GO:0010871 |
negative regulation of receptor biosynthetic process(GO:0010871) |
| 0.1 |
0.4 |
GO:0006189 |
'de novo' IMP biosynthetic process(GO:0006189) |
| 0.1 |
0.2 |
GO:1990046 |
positive regulation of mitochondrial DNA replication(GO:0090297) regulation of cardiolipin metabolic process(GO:1900208) positive regulation of cardiolipin metabolic process(GO:1900210) stress-induced mitochondrial fusion(GO:1990046) |
| 0.1 |
0.5 |
GO:0002634 |
regulation of germinal center formation(GO:0002634) |
| 0.1 |
0.3 |
GO:0002337 |
B-1a B cell differentiation(GO:0002337) |
| 0.1 |
0.1 |
GO:0048633 |
positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.1 |
0.7 |
GO:0035871 |
protein K11-linked deubiquitination(GO:0035871) |
| 0.1 |
0.7 |
GO:1901250 |
regulation of lung goblet cell differentiation(GO:1901249) negative regulation of lung goblet cell differentiation(GO:1901250) |
| 0.1 |
0.7 |
GO:0070131 |
positive regulation of mitochondrial translation(GO:0070131) |
| 0.1 |
0.3 |
GO:0060754 |
positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.1 |
0.8 |
GO:0060019 |
radial glial cell differentiation(GO:0060019) |
| 0.1 |
0.2 |
GO:0090399 |
replicative senescence(GO:0090399) |
| 0.1 |
1.3 |
GO:0099517 |
anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.1 |
0.9 |
GO:0038063 |
collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.1 |
0.3 |
GO:0040031 |
snRNA modification(GO:0040031) |
| 0.1 |
0.3 |
GO:0016259 |
selenocysteine metabolic process(GO:0016259) |
| 0.1 |
1.0 |
GO:0007530 |
sex determination(GO:0007530) |
| 0.1 |
0.2 |
GO:0045903 |
positive regulation of translational fidelity(GO:0045903) |
| 0.1 |
0.1 |
GO:0071680 |
response to indole-3-methanol(GO:0071680) cellular response to indole-3-methanol(GO:0071681) |
| 0.1 |
0.3 |
GO:0009052 |
pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.1 |
1.1 |
GO:0097284 |
hepatocyte apoptotic process(GO:0097284) |
| 0.1 |
0.8 |
GO:0006388 |
tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.1 |
0.4 |
GO:0051639 |
actin filament network formation(GO:0051639) |
| 0.1 |
0.2 |
GO:0006407 |
rRNA export from nucleus(GO:0006407) |
| 0.1 |
1.7 |
GO:0043153 |
entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.1 |
0.3 |
GO:0070474 |
positive regulation of uterine smooth muscle contraction(GO:0070474) |
| 0.1 |
0.5 |
GO:0018401 |
peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.1 |
0.3 |
GO:0010273 |
detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.1 |
0.2 |
GO:0036388 |
pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.1 |
0.4 |
GO:0001842 |
neural fold formation(GO:0001842) |
| 0.1 |
0.3 |
GO:0000160 |
phosphorelay signal transduction system(GO:0000160) |
| 0.1 |
0.2 |
GO:1990928 |
response to amino acid starvation(GO:1990928) |
| 0.1 |
0.1 |
GO:0048630 |
skeletal muscle tissue growth(GO:0048630) |
| 0.1 |
0.2 |
GO:0019530 |
taurine metabolic process(GO:0019530) |
| 0.1 |
0.3 |
GO:1903715 |
regulation of aerobic respiration(GO:1903715) |
| 0.1 |
0.1 |
GO:1900377 |
negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.1 |
1.2 |
GO:2000060 |
positive regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000060) |
| 0.1 |
0.1 |
GO:0048208 |
vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.1 |
0.3 |
GO:0097680 |
double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.1 |
1.1 |
GO:0035873 |
lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.1 |
0.2 |
GO:0015888 |
thiamine transport(GO:0015888) |
| 0.1 |
0.2 |
GO:0006529 |
asparagine biosynthetic process(GO:0006529) |
| 0.1 |
0.1 |
GO:0046121 |
deoxyribonucleoside catabolic process(GO:0046121) |
| 0.1 |
0.4 |
GO:0033148 |
positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.1 |
0.3 |
GO:0043985 |
histone H4-R3 methylation(GO:0043985) |
| 0.1 |
0.4 |
GO:0044351 |
macropinocytosis(GO:0044351) |
| 0.1 |
2.0 |
GO:0048255 |
mRNA stabilization(GO:0048255) |
| 0.0 |
0.5 |
GO:0046036 |
GTP biosynthetic process(GO:0006183) CTP biosynthetic process(GO:0006241) CTP metabolic process(GO:0046036) |
| 0.0 |
0.3 |
GO:1904417 |
regulation of xenophagy(GO:1904415) positive regulation of xenophagy(GO:1904417) |
| 0.0 |
0.3 |
GO:0032483 |
regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 |
0.2 |
GO:1901970 |
positive regulation of mitotic sister chromatid separation(GO:1901970) |
| 0.0 |
0.7 |
GO:0007035 |
vacuolar acidification(GO:0007035) |
| 0.0 |
0.0 |
GO:1900369 |
negative regulation of RNA interference(GO:1900369) |
| 0.0 |
0.6 |
GO:0001731 |
formation of translation preinitiation complex(GO:0001731) |
| 0.0 |
0.2 |
GO:0090220 |
meiotic telomere clustering(GO:0045141) chromosome localization to nuclear envelope involved in homologous chromosome segregation(GO:0090220) |
| 0.0 |
0.1 |
GO:0016095 |
polyprenol catabolic process(GO:0016095) |
| 0.0 |
0.1 |
GO:0097296 |
activation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097296) |
| 0.0 |
0.2 |
GO:0070129 |
regulation of mitochondrial translation(GO:0070129) |
| 0.0 |
0.4 |
GO:0030174 |
regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.0 |
0.3 |
GO:0090557 |
establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 |
0.0 |
GO:0051595 |
response to methylglyoxal(GO:0051595) |
| 0.0 |
0.4 |
GO:0032836 |
glomerular basement membrane development(GO:0032836) |
| 0.0 |
0.1 |
GO:0060482 |
lobar bronchus development(GO:0060482) |
| 0.0 |
0.3 |
GO:0042590 |
antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.0 |
0.2 |
GO:0045607 |
regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 |
0.2 |
GO:0046726 |
positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.0 |
0.1 |
GO:0015938 |
coenzyme A catabolic process(GO:0015938) nucleoside bisphosphate catabolic process(GO:0033869) ribonucleoside bisphosphate catabolic process(GO:0034031) purine nucleoside bisphosphate catabolic process(GO:0034034) |
| 0.0 |
0.2 |
GO:0007171 |
activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 |
0.1 |
GO:0006421 |
asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 |
0.0 |
GO:0045583 |
regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 0.0 |
0.2 |
GO:0036324 |
vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 0.0 |
0.2 |
GO:0035331 |
negative regulation of hippo signaling(GO:0035331) |
| 0.0 |
0.2 |
GO:1904694 |
negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 |
0.1 |
GO:0035878 |
nail development(GO:0035878) |
| 0.0 |
0.2 |
GO:0048304 |
positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.0 |
0.2 |
GO:0060837 |
blood vessel endothelial cell differentiation(GO:0060837) |
| 0.0 |
1.3 |
GO:0048011 |
neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.0 |
1.3 |
GO:0008608 |
attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.0 |
0.6 |
GO:0051974 |
negative regulation of telomerase activity(GO:0051974) |
| 0.0 |
0.4 |
GO:0003356 |
regulation of cilium movement(GO:0003352) regulation of cilium beat frequency(GO:0003356) |
| 0.0 |
0.1 |
GO:0043456 |
regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 |
0.1 |
GO:0002014 |
vasoconstriction of artery involved in ischemic response to lowering of systemic arterial blood pressure(GO:0002014) |
| 0.0 |
0.0 |
GO:0070428 |
nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) |
| 0.0 |
0.5 |
GO:0006817 |
phosphate ion transport(GO:0006817) |
| 0.0 |
0.1 |
GO:0072610 |
interleukin-12 secretion(GO:0072610) regulation of interleukin-12 secretion(GO:2001182) positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.0 |
0.3 |
GO:0035635 |
entry of bacterium into host cell(GO:0035635) regulation of entry of bacterium into host cell(GO:2000535) |
| 0.0 |
0.1 |
GO:0090271 |
positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.0 |
0.2 |
GO:0006265 |
DNA topological change(GO:0006265) |
| 0.0 |
0.2 |
GO:0090073 |
positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 |
0.0 |
GO:1902262 |
apoptotic process involved in patterning of blood vessels(GO:1902262) |
| 0.0 |
0.1 |
GO:0051866 |
general adaptation syndrome(GO:0051866) |
| 0.0 |
0.1 |
GO:0050748 |
negative regulation of lipoprotein metabolic process(GO:0050748) |
| 0.0 |
0.5 |
GO:0006268 |
DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 |
0.3 |
GO:0071364 |
response to epidermal growth factor(GO:0070849) cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.0 |
0.1 |
GO:0046294 |
formaldehyde catabolic process(GO:0046294) |
| 0.0 |
0.2 |
GO:1903347 |
negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.0 |
0.0 |
GO:0033278 |
cell proliferation in midbrain(GO:0033278) |
| 0.0 |
0.2 |
GO:0014886 |
transition between slow and fast fiber(GO:0014886) |
| 0.0 |
0.1 |
GO:1902358 |
sulfate transmembrane transport(GO:1902358) |
| 0.0 |
0.3 |
GO:0045719 |
negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 |
0.1 |
GO:0060056 |
mammary gland involution(GO:0060056) |
| 0.0 |
0.1 |
GO:0015882 |
L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.0 |
0.4 |
GO:0071803 |
positive regulation of podosome assembly(GO:0071803) |
| 0.0 |
0.7 |
GO:0042438 |
melanin biosynthetic process(GO:0042438) |
| 0.0 |
0.0 |
GO:0006983 |
ER overload response(GO:0006983) |
| 0.0 |
0.1 |
GO:0033326 |
cerebrospinal fluid secretion(GO:0033326) |
| 0.0 |
0.3 |
GO:0035520 |
monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 |
0.1 |
GO:0000447 |
endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.0 |
0.9 |
GO:0010972 |
negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.0 |
0.1 |
GO:0060416 |
response to growth hormone(GO:0060416) |
| 0.0 |
1.8 |
GO:0072661 |
protein targeting to plasma membrane(GO:0072661) |
| 0.0 |
0.2 |
GO:0000480 |
endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 |
0.2 |
GO:0061084 |
regulation of protein refolding(GO:0061083) negative regulation of protein refolding(GO:0061084) |
| 0.0 |
0.1 |
GO:0009644 |
response to high light intensity(GO:0009644) |
| 0.0 |
0.2 |
GO:0010917 |
negative regulation of mitochondrial membrane potential(GO:0010917) |
| 0.0 |
0.2 |
GO:0006123 |
mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 |
0.4 |
GO:1990403 |
embryonic brain development(GO:1990403) |
| 0.0 |
0.2 |
GO:1904217 |
regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) regulation of serine C-palmitoyltransferase activity(GO:1904220) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.0 |
2.5 |
GO:0006405 |
RNA export from nucleus(GO:0006405) |
| 0.0 |
0.2 |
GO:0071285 |
cellular response to lithium ion(GO:0071285) |
| 0.0 |
0.1 |
GO:0042518 |
negative regulation of tyrosine phosphorylation of Stat3 protein(GO:0042518) |
| 0.0 |
0.2 |
GO:0003222 |
ventricular trabecula myocardium morphogenesis(GO:0003222) |
| 0.0 |
0.2 |
GO:0034587 |
piRNA metabolic process(GO:0034587) |
| 0.0 |
0.3 |
GO:0030388 |
fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 |
0.2 |
GO:0090435 |
protein localization to nuclear envelope(GO:0090435) |
| 0.0 |
1.0 |
GO:0006101 |
citrate metabolic process(GO:0006101) |
| 0.0 |
0.2 |
GO:0045948 |
positive regulation of translational initiation(GO:0045948) |
| 0.0 |
0.2 |
GO:0098903 |
regulation of membrane repolarization during action potential(GO:0098903) |
| 0.0 |
0.1 |
GO:0050812 |
regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.0 |
0.1 |
GO:1990379 |
lipid transport across blood brain barrier(GO:1990379) |
| 0.0 |
0.0 |
GO:1902745 |
positive regulation of lamellipodium assembly(GO:0010592) positive regulation of lamellipodium organization(GO:1902745) |
| 0.0 |
0.1 |
GO:1902512 |
positive regulation of apoptotic DNA fragmentation(GO:1902512) positive regulation of DNA catabolic process(GO:1903626) |
| 0.0 |
0.1 |
GO:2000348 |
regulation of CD40 signaling pathway(GO:2000348) |
| 0.0 |
0.2 |
GO:0006177 |
GMP biosynthetic process(GO:0006177) |
| 0.0 |
0.0 |
GO:0045213 |
neurotransmitter receptor metabolic process(GO:0045213) |
| 0.0 |
0.1 |
GO:0006438 |
valyl-tRNA aminoacylation(GO:0006438) |
| 0.0 |
0.1 |
GO:0010808 |
positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.0 |
0.1 |
GO:2000383 |
regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.0 |
0.1 |
GO:0097212 |
late endosomal microautophagy(GO:0061738) lysosomal membrane organization(GO:0097212) |
| 0.0 |
0.7 |
GO:0032008 |
positive regulation of TOR signaling(GO:0032008) |
| 0.0 |
0.2 |
GO:0008616 |
queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 |
0.9 |
GO:0010501 |
RNA secondary structure unwinding(GO:0010501) |
| 0.0 |
0.4 |
GO:0021542 |
dentate gyrus development(GO:0021542) |
| 0.0 |
0.2 |
GO:0045603 |
positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 |
0.1 |
GO:0008612 |
peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.0 |
0.3 |
GO:0019227 |
neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 |
0.8 |
GO:0000462 |
maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 |
0.3 |
GO:0080111 |
DNA demethylation(GO:0080111) |
| 0.0 |
0.2 |
GO:0036006 |
response to macrophage colony-stimulating factor(GO:0036005) cellular response to macrophage colony-stimulating factor stimulus(GO:0036006) |
| 0.0 |
1.8 |
GO:0043966 |
histone H3 acetylation(GO:0043966) |
| 0.0 |
0.4 |
GO:0009048 |
dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 |
0.3 |
GO:0050849 |
negative regulation of calcium-mediated signaling(GO:0050849) |
| 0.0 |
0.1 |
GO:2000525 |
regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.0 |
0.0 |
GO:0042726 |
flavin-containing compound metabolic process(GO:0042726) |
| 0.0 |
0.1 |
GO:0070782 |
phosphatidylserine exposure on apoptotic cell surface(GO:0070782) regulation of potassium ion export(GO:1902302) |
| 0.0 |
0.1 |
GO:1903232 |
melanosome assembly(GO:1903232) |
| 0.0 |
0.1 |
GO:1902993 |
positive regulation of beta-amyloid formation(GO:1902004) positive regulation of amyloid precursor protein catabolic process(GO:1902993) |
| 0.0 |
0.6 |
GO:0009303 |
rRNA transcription(GO:0009303) |
| 0.0 |
0.1 |
GO:0036518 |
chemorepulsion of dopaminergic neuron axon(GO:0036518) |
| 0.0 |
0.2 |
GO:0036444 |
calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 |
0.2 |
GO:0010807 |
regulation of synaptic vesicle priming(GO:0010807) |
| 0.0 |
0.6 |
GO:0035329 |
hippo signaling(GO:0035329) |
| 0.0 |
0.2 |
GO:0003433 |
chondrocyte development involved in endochondral bone morphogenesis(GO:0003433) |
| 0.0 |
0.1 |
GO:0009125 |
nucleoside monophosphate catabolic process(GO:0009125) |
| 0.0 |
0.2 |
GO:0003338 |
metanephros morphogenesis(GO:0003338) |
| 0.0 |
0.1 |
GO:0000414 |
regulation of histone H3-K36 methylation(GO:0000414) |
| 0.0 |
0.0 |
GO:0046084 |
adenine metabolic process(GO:0046083) adenine biosynthetic process(GO:0046084) |
| 0.0 |
0.3 |
GO:0034122 |
negative regulation of toll-like receptor signaling pathway(GO:0034122) |
| 0.0 |
0.1 |
GO:0032328 |
alanine transport(GO:0032328) |
| 0.0 |
0.1 |
GO:0035063 |
nuclear speck organization(GO:0035063) |
| 0.0 |
0.2 |
GO:0070307 |
lens fiber cell development(GO:0070307) |
| 0.0 |
0.2 |
GO:0031536 |
positive regulation of exit from mitosis(GO:0031536) |
| 0.0 |
0.2 |
GO:0090232 |
positive regulation of spindle checkpoint(GO:0090232) |
| 0.0 |
0.0 |
GO:0006348 |
chromatin silencing at telomere(GO:0006348) |
| 0.0 |
0.1 |
GO:0043553 |
negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 |
0.4 |
GO:0038092 |
nodal signaling pathway(GO:0038092) |
| 0.0 |
0.2 |
GO:1903298 |
regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903297) negative regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903298) intrinsic apoptotic signaling pathway in response to hypoxia(GO:1990144) |
| 0.0 |
0.1 |
GO:1900275 |
B cell proliferation involved in immune response(GO:0002322) negative regulation of phospholipase C activity(GO:1900275) |
| 0.0 |
0.1 |
GO:0040016 |
embryonic cleavage(GO:0040016) |
| 0.0 |
0.1 |
GO:0001978 |
regulation of systemic arterial blood pressure by carotid sinus baroreceptor feedback(GO:0001978) |
| 0.0 |
0.3 |
GO:0002385 |
mucosal immune response(GO:0002385) |
| 0.0 |
0.0 |
GO:0071649 |
regulation of chemokine (C-C motif) ligand 5 production(GO:0071649) |
| 0.0 |
0.1 |
GO:0045716 |
positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.0 |
0.0 |
GO:0071472 |
cellular response to salt stress(GO:0071472) |
| 0.0 |
0.2 |
GO:0006020 |
inositol metabolic process(GO:0006020) |
| 0.0 |
1.7 |
GO:0032526 |
response to retinoic acid(GO:0032526) |
| 0.0 |
0.1 |
GO:0034227 |
tRNA thio-modification(GO:0034227) |
| 0.0 |
0.1 |
GO:0097278 |
virion attachment to host cell(GO:0019062) adhesion of symbiont to host cell(GO:0044650) complement-dependent cytotoxicity(GO:0097278) |
| 0.0 |
0.1 |
GO:0060468 |
prevention of polyspermy(GO:0060468) |
| 0.0 |
0.0 |
GO:1902608 |
regulation of large conductance calcium-activated potassium channel activity(GO:1902606) positive regulation of large conductance calcium-activated potassium channel activity(GO:1902608) |
| 0.0 |
0.5 |
GO:0046856 |
phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 |
0.3 |
GO:0031340 |
positive regulation of vesicle fusion(GO:0031340) |
| 0.0 |
0.0 |
GO:0000479 |
endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000479) |
| 0.0 |
0.1 |
GO:0006668 |
sphinganine-1-phosphate metabolic process(GO:0006668) |
| 0.0 |
0.1 |
GO:0000727 |
double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 |
0.2 |
GO:2000489 |
hepatic stellate cell activation(GO:0035733) regulation of hepatic stellate cell activation(GO:2000489) |
| 0.0 |
0.0 |
GO:0042256 |
mature ribosome assembly(GO:0042256) |
| 0.0 |
0.1 |
GO:1900220 |
semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.0 |
0.5 |
GO:0042149 |
cellular response to glucose starvation(GO:0042149) |
| 0.0 |
0.1 |
GO:0051694 |
pointed-end actin filament capping(GO:0051694) |
| 0.0 |
0.1 |
GO:0045351 |
type I interferon biosynthetic process(GO:0045351) |
| 0.0 |
0.2 |
GO:0070886 |
positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 |
0.1 |
GO:0006003 |
fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 |
0.0 |
GO:0048318 |
axial mesoderm development(GO:0048318) |
| 0.0 |
0.1 |
GO:2000786 |
positive regulation of autophagosome assembly(GO:2000786) |
| 0.0 |
0.0 |
GO:0032286 |
central nervous system myelin maintenance(GO:0032286) |
| 0.0 |
0.1 |
GO:0090035 |
regulation of chaperone-mediated protein complex assembly(GO:0090034) positive regulation of chaperone-mediated protein complex assembly(GO:0090035) |
| 0.0 |
0.2 |
GO:0023021 |
termination of signal transduction(GO:0023021) |
| 0.0 |
0.1 |
GO:0032485 |
regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 |
0.2 |
GO:0000244 |
spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 |
0.3 |
GO:0032753 |
positive regulation of interleukin-4 production(GO:0032753) |
| 0.0 |
0.1 |
GO:1900747 |
negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 |
0.1 |
GO:0015870 |
acetylcholine transport(GO:0015870) acetate ester transport(GO:1901374) |
| 0.0 |
0.1 |
GO:0009098 |
branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.0 |
0.6 |
GO:0018208 |
peptidyl-proline modification(GO:0018208) |
| 0.0 |
0.3 |
GO:0043248 |
proteasome assembly(GO:0043248) |
| 0.0 |
0.0 |
GO:1904338 |
regulation of dopaminergic neuron differentiation(GO:1904338) |
| 0.0 |
0.1 |
GO:0051189 |
Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 |
0.3 |
GO:0048026 |
positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 |
0.1 |
GO:0019919 |
peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) peptidyl-arginine N-methylation(GO:0035246) peptidyl-arginine omega-N-methylation(GO:0035247) |
| 0.0 |
0.1 |
GO:0015786 |
UDP-glucose transport(GO:0015786) |
| 0.0 |
0.2 |
GO:0043162 |
ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 |
0.1 |
GO:0050901 |
leukocyte tethering or rolling(GO:0050901) |
| 0.0 |
0.1 |
GO:1903300 |
negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 |
0.5 |
GO:0015991 |
energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.0 |
0.1 |
GO:0033572 |
transferrin transport(GO:0033572) |
| 0.0 |
0.3 |
GO:0006054 |
N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 |
0.1 |
GO:0007603 |
phototransduction, visible light(GO:0007603) |
| 0.0 |
0.1 |
GO:2000767 |
positive regulation of cytoplasmic translation(GO:2000767) |
| 0.0 |
0.3 |
GO:0060669 |
embryonic placenta morphogenesis(GO:0060669) |
| 0.0 |
0.1 |
GO:2000172 |
regulation of branching morphogenesis of a nerve(GO:2000172) |
| 0.0 |
0.1 |
GO:0044027 |
hypermethylation of CpG island(GO:0044027) |
| 0.0 |
0.6 |
GO:0007157 |
heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 |
0.1 |
GO:0046061 |
dATP catabolic process(GO:0046061) |
| 0.0 |
0.1 |
GO:0071044 |
histone mRNA catabolic process(GO:0071044) |
| 0.0 |
0.3 |
GO:0042407 |
cristae formation(GO:0042407) |
| 0.0 |
0.1 |
GO:0035606 |
peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.0 |
0.5 |
GO:0045104 |
intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 |
0.7 |
GO:0071479 |
cellular response to ionizing radiation(GO:0071479) |
| 0.0 |
0.2 |
GO:0019368 |
fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 |
0.3 |
GO:0030213 |
hyaluronan biosynthetic process(GO:0030213) |
| 0.0 |
0.2 |
GO:0035414 |
negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 |
0.0 |
GO:0097211 |
response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.0 |
0.0 |
GO:0051204 |
protein insertion into mitochondrial membrane(GO:0051204) |
| 0.0 |
0.3 |
GO:0050650 |
chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.0 |
0.7 |
GO:0016925 |
protein sumoylation(GO:0016925) |
| 0.0 |
0.0 |
GO:0019087 |
transformation of host cell by virus(GO:0019087) |
| 0.0 |
0.1 |
GO:0070966 |
nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 |
0.1 |
GO:1990776 |
cellular response to angiotensin(GO:1904385) response to angiotensin(GO:1990776) |
| 0.0 |
0.2 |
GO:0032780 |
negative regulation of ATPase activity(GO:0032780) |
| 0.0 |
0.1 |
GO:0009235 |
cobalamin metabolic process(GO:0009235) |
| 0.0 |
0.1 |
GO:0002566 |
somatic diversification of immune receptors via somatic mutation(GO:0002566) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 |
0.1 |
GO:0010667 |
negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.0 |
0.0 |
GO:0033615 |
mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.0 |
0.2 |
GO:2000741 |
positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.0 |
0.1 |
GO:1902035 |
positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.0 |
0.1 |
GO:0007168 |
receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 |
0.1 |
GO:0061052 |
negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 0.0 |
0.1 |
GO:0006004 |
fucose metabolic process(GO:0006004) |
| 0.0 |
0.1 |
GO:0006287 |
base-excision repair, gap-filling(GO:0006287) |
| 0.0 |
0.3 |
GO:0006446 |
regulation of translational initiation(GO:0006446) |
| 0.0 |
0.0 |
GO:0042492 |
gamma-delta T cell differentiation(GO:0042492) regulation of gamma-delta T cell differentiation(GO:0045586) gamma-delta T cell activation(GO:0046629) regulation of gamma-delta T cell activation(GO:0046643) |
| 0.0 |
0.3 |
GO:0000387 |
spliceosomal snRNP assembly(GO:0000387) |
| 0.0 |
0.7 |
GO:0035914 |
skeletal muscle cell differentiation(GO:0035914) |
| 0.0 |
0.1 |
GO:0090148 |
membrane fission(GO:0090148) |
| 0.0 |
0.3 |
GO:0070584 |
mitochondrion morphogenesis(GO:0070584) |
| 0.0 |
0.1 |
GO:0034498 |
early endosome to Golgi transport(GO:0034498) |
| 0.0 |
0.8 |
GO:0051865 |
protein autoubiquitination(GO:0051865) |
| 0.0 |
0.0 |
GO:0030327 |
sulfur amino acid catabolic process(GO:0000098) prenylated protein catabolic process(GO:0030327) |
| 0.0 |
0.0 |
GO:1903800 |
positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 |
0.1 |
GO:0060765 |
regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.0 |
0.1 |
GO:0030263 |
apoptotic chromosome condensation(GO:0030263) |
| 0.0 |
0.1 |
GO:0045072 |
regulation of interferon-gamma biosynthetic process(GO:0045072) positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.0 |
0.0 |
GO:0008065 |
establishment of blood-nerve barrier(GO:0008065) |
| 0.0 |
0.1 |
GO:0000028 |
ribosomal small subunit assembly(GO:0000028) |
| 0.0 |
0.1 |
GO:0006957 |
complement activation, alternative pathway(GO:0006957) |
| 0.0 |
0.2 |
GO:0030574 |
collagen catabolic process(GO:0030574) |
| 0.0 |
0.0 |
GO:1902626 |
assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 |
0.1 |
GO:1990173 |
regulation of establishment of protein localization to chromosome(GO:0070202) regulation of establishment of protein localization to telomere(GO:0070203) protein localization to nuclear body(GO:1903405) regulation of protein localization to chromosome, telomeric region(GO:1904814) positive regulation of protein localization to chromosome, telomeric region(GO:1904816) positive regulation of establishment of protein localization to telomere(GO:1904851) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) protein localization to nucleoplasm(GO:1990173) |
| 0.0 |
0.1 |
GO:0051611 |
negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 |
0.4 |
GO:0032728 |
positive regulation of interferon-beta production(GO:0032728) |
| 0.0 |
0.1 |
GO:2000279 |
negative regulation of DNA biosynthetic process(GO:2000279) |
| 0.0 |
0.2 |
GO:0014823 |
response to activity(GO:0014823) |
| 0.0 |
0.2 |
GO:0014741 |
negative regulation of cardiac muscle hypertrophy(GO:0010614) negative regulation of muscle hypertrophy(GO:0014741) |
| 0.0 |
0.3 |
GO:0048488 |
synaptic vesicle endocytosis(GO:0048488) |
| 0.0 |
0.0 |
GO:0060346 |
bone trabecula formation(GO:0060346) |
| 0.0 |
0.1 |
GO:0006977 |
DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.0 |
0.1 |
GO:0031509 |
telomeric heterochromatin assembly(GO:0031509) negative regulation of chromosome condensation(GO:1902340) |
| 0.0 |
0.1 |
GO:1902414 |
protein localization to cell junction(GO:1902414) |
| 0.0 |
0.1 |
GO:0001504 |
neurotransmitter uptake(GO:0001504) |
| 0.0 |
0.0 |
GO:0051490 |
negative regulation of filopodium assembly(GO:0051490) |
| 0.0 |
0.1 |
GO:0090261 |
positive regulation of inclusion body assembly(GO:0090261) |
| 0.0 |
0.2 |
GO:0000578 |
embryonic axis specification(GO:0000578) |
| 0.0 |
0.5 |
GO:0015992 |
proton transport(GO:0015992) |
| 0.0 |
0.0 |
GO:0010881 |
regulation of cardiac muscle contraction by regulation of the release of sequestered calcium ion(GO:0010881) |
| 0.0 |
0.2 |
GO:0048025 |
negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 |
0.1 |
GO:0048671 |
negative regulation of collateral sprouting(GO:0048671) |
| 0.0 |
0.1 |
GO:1904321 |
response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.0 |
0.6 |
GO:0051453 |
regulation of intracellular pH(GO:0051453) |
| 0.0 |
0.2 |
GO:0015804 |
neutral amino acid transport(GO:0015804) |
| 0.0 |
0.1 |
GO:0021684 |
cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 |
0.2 |
GO:0030318 |
melanocyte differentiation(GO:0030318) |
| 0.0 |
0.0 |
GO:0043383 |
negative T cell selection(GO:0043383) negative thymic T cell selection(GO:0045060) |
| 0.0 |
0.4 |
GO:0048747 |
muscle fiber development(GO:0048747) |
| 0.0 |
0.0 |
GO:1900016 |
negative regulation of cytokine production involved in inflammatory response(GO:1900016) |
| 0.0 |
0.2 |
GO:0030513 |
positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 |
0.0 |
GO:0048496 |
maintenance of organ identity(GO:0048496) |
| 0.0 |
0.0 |
GO:0010958 |
regulation of amino acid import(GO:0010958) regulation of L-arginine import(GO:0010963) |
| 0.0 |
0.3 |
GO:0043550 |
regulation of lipid kinase activity(GO:0043550) |
| 0.0 |
0.0 |
GO:0000715 |
nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.0 |
0.2 |
GO:0032933 |
response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 |
0.1 |
GO:0035994 |
response to muscle stretch(GO:0035994) |
| 0.0 |
0.1 |
GO:0035563 |
positive regulation of chromatin binding(GO:0035563) |
| 0.0 |
0.0 |
GO:0000429 |
carbon catabolite regulation of transcription from RNA polymerase II promoter(GO:0000429) regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) carbon catabolite activation of transcription from RNA polymerase II promoter(GO:0000436) |
| 0.0 |
0.0 |
GO:0009298 |
GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 |
0.1 |
GO:0044381 |
glucose import in response to insulin stimulus(GO:0044381) regulation of glucose import in response to insulin stimulus(GO:2001273) |
| 0.0 |
0.2 |
GO:1902230 |
negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.0 |
0.1 |
GO:2000394 |
positive regulation of lamellipodium morphogenesis(GO:2000394) |
| 0.0 |
0.1 |
GO:0045717 |
negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.0 |
0.0 |
GO:0032272 |
negative regulation of protein polymerization(GO:0032272) |
| 0.0 |
0.5 |
GO:0007405 |
neuroblast proliferation(GO:0007405) |
| 0.0 |
0.1 |
GO:0002689 |
negative regulation of leukocyte chemotaxis(GO:0002689) |
| 0.0 |
0.2 |
GO:0032981 |
NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 |
0.1 |
GO:2000623 |
regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 |
0.1 |
GO:0019585 |
uronic acid metabolic process(GO:0006063) glucuronate metabolic process(GO:0019585) cellular glucuronidation(GO:0052695) |
| 0.0 |
0.0 |
GO:1904008 |
cellular response to salt(GO:1902075) response to monosodium glutamate(GO:1904008) cellular response to monosodium glutamate(GO:1904009) |
| 0.0 |
0.0 |
GO:0031086 |
nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 |
0.0 |
GO:1904058 |
regulation of gastric acid secretion(GO:0060453) positive regulation of sensory perception of pain(GO:1904058) |
| 0.0 |
0.1 |
GO:0032402 |
melanosome transport(GO:0032402) |
| 0.0 |
0.1 |
GO:0050853 |
B cell receptor signaling pathway(GO:0050853) |
| 0.0 |
0.4 |
GO:0032543 |
mitochondrial translation(GO:0032543) |
| 0.0 |
0.1 |
GO:1901339 |
regulation of store-operated calcium channel activity(GO:1901339) |
| 0.0 |
0.1 |
GO:0042769 |
DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 |
0.0 |
GO:1902916 |
positive regulation of protein polyubiquitination(GO:1902916) |
| 0.0 |
0.1 |
GO:0006298 |
mismatch repair(GO:0006298) |
| 0.0 |
0.0 |
GO:0051031 |
tRNA transport(GO:0051031) |
| 0.0 |
0.2 |
GO:0016226 |
iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 |
0.2 |
GO:0030488 |
tRNA methylation(GO:0030488) |
| 0.0 |
0.1 |
GO:0036506 |
maintenance of unfolded protein(GO:0036506) protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.0 |
0.0 |
GO:0070164 |
negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 |
0.1 |
GO:0034379 |
very-low-density lipoprotein particle assembly(GO:0034379) |