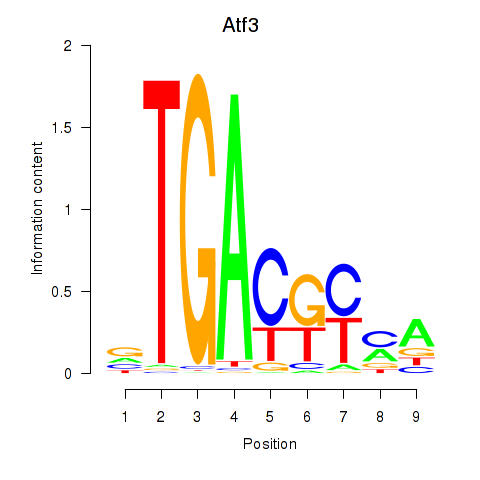
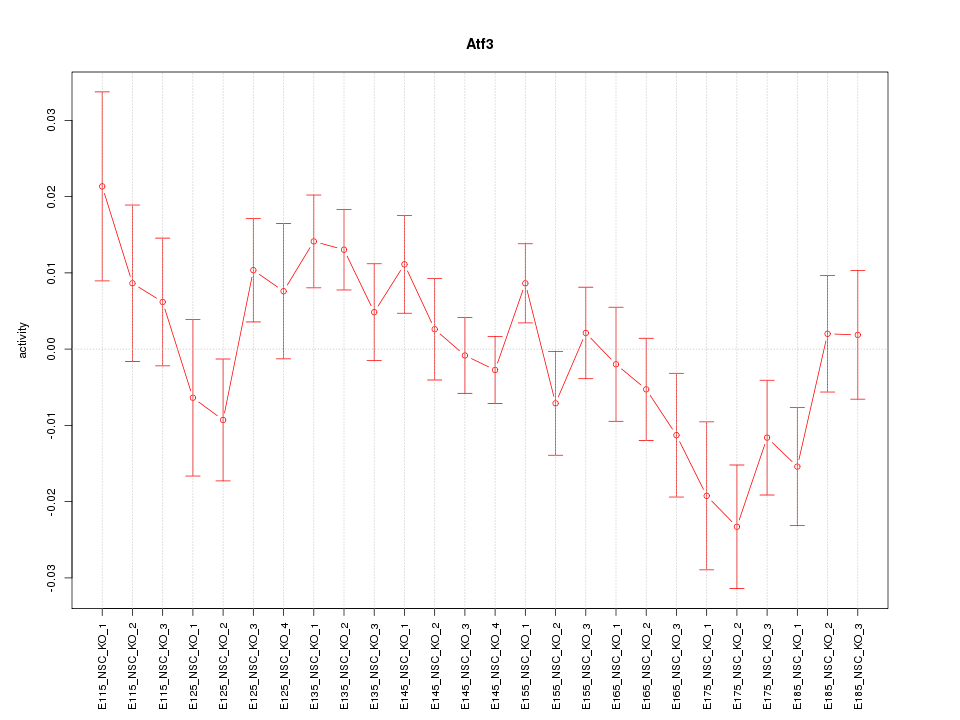
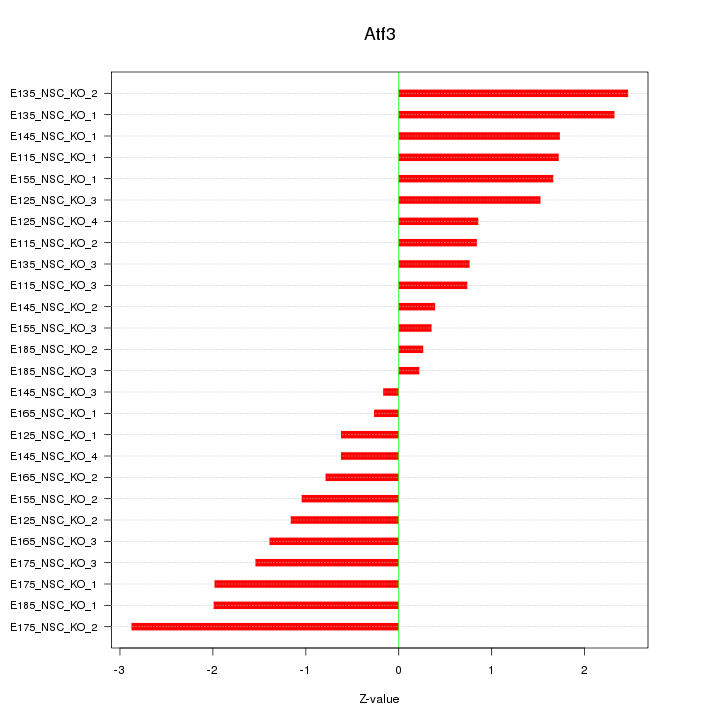
| 1.4 |
4.3 |
GO:0060598 |
dichotomous subdivision of terminal units involved in mammary gland duct morphogenesis(GO:0060598) |
| 1.0 |
3.0 |
GO:0038203 |
TORC2 signaling(GO:0038203) |
| 0.9 |
2.7 |
GO:0036388 |
pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.9 |
4.4 |
GO:0051315 |
attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.8 |
2.5 |
GO:0031120 |
snRNA pseudouridine synthesis(GO:0031120) |
| 0.8 |
3.1 |
GO:1900145 |
regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900175) positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.8 |
4.5 |
GO:0021797 |
forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.6 |
1.9 |
GO:0071921 |
establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.6 |
1.9 |
GO:1904274 |
tricellular tight junction assembly(GO:1904274) |
| 0.6 |
5.5 |
GO:1904668 |
positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.6 |
2.9 |
GO:0030951 |
establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.6 |
2.9 |
GO:0046601 |
positive regulation of centriole replication(GO:0046601) |
| 0.6 |
2.2 |
GO:0072385 |
minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.5 |
1.6 |
GO:0070634 |
transepithelial ammonium transport(GO:0070634) |
| 0.5 |
1.1 |
GO:1905065 |
positive regulation of vascular smooth muscle cell differentiation(GO:1905065) |
| 0.5 |
1.6 |
GO:0006931 |
substrate-dependent cell migration, cell attachment to substrate(GO:0006931) calcium-independent cell-matrix adhesion(GO:0007161) |
| 0.5 |
1.6 |
GO:0072139 |
glomerular parietal epithelial cell differentiation(GO:0072139) |
| 0.5 |
1.5 |
GO:1901552 |
positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.5 |
4.3 |
GO:0002318 |
myeloid progenitor cell differentiation(GO:0002318) |
| 0.5 |
1.4 |
GO:0002302 |
CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 0.5 |
1.9 |
GO:0070318 |
positive regulation of G0 to G1 transition(GO:0070318) |
| 0.5 |
1.4 |
GO:0030167 |
proteoglycan catabolic process(GO:0030167) |
| 0.5 |
1.4 |
GO:0048254 |
snoRNA localization(GO:0048254) |
| 0.5 |
2.7 |
GO:0009786 |
regulation of asymmetric cell division(GO:0009786) |
| 0.4 |
1.8 |
GO:0046836 |
glycolipid transport(GO:0046836) |
| 0.4 |
0.9 |
GO:0006059 |
hexitol metabolic process(GO:0006059) |
| 0.4 |
0.9 |
GO:0006610 |
ribosomal protein import into nucleus(GO:0006610) |
| 0.4 |
1.3 |
GO:0006083 |
acetate metabolic process(GO:0006083) |
| 0.4 |
0.4 |
GO:0070212 |
protein poly-ADP-ribosylation(GO:0070212) |
| 0.4 |
2.6 |
GO:0046602 |
regulation of mitotic centrosome separation(GO:0046602) |
| 0.4 |
1.3 |
GO:0046381 |
CMP-N-acetylneuraminate metabolic process(GO:0046381) |
| 0.4 |
1.7 |
GO:0070829 |
heterochromatin maintenance(GO:0070829) |
| 0.4 |
2.0 |
GO:0060051 |
negative regulation of protein glycosylation(GO:0060051) |
| 0.4 |
5.6 |
GO:0034501 |
protein localization to kinetochore(GO:0034501) |
| 0.4 |
1.6 |
GO:1904565 |
response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.4 |
1.6 |
GO:0045617 |
negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.4 |
2.7 |
GO:0042590 |
antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.4 |
1.1 |
GO:0001978 |
regulation of systemic arterial blood pressure by carotid sinus baroreceptor feedback(GO:0001978) |
| 0.4 |
1.5 |
GO:0021698 |
cerebellar cortex structural organization(GO:0021698) |
| 0.4 |
1.5 |
GO:0014722 |
regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.4 |
2.3 |
GO:1902410 |
mitotic cytokinetic process(GO:1902410) |
| 0.4 |
0.7 |
GO:0048296 |
isotype switching to IgA isotypes(GO:0048290) regulation of isotype switching to IgA isotypes(GO:0048296) |
| 0.4 |
1.1 |
GO:0009106 |
lipoate metabolic process(GO:0009106) |
| 0.4 |
1.8 |
GO:0015671 |
oxygen transport(GO:0015671) |
| 0.4 |
1.4 |
GO:0002949 |
tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.4 |
1.1 |
GO:0046370 |
fructose biosynthetic process(GO:0046370) |
| 0.4 |
1.8 |
GO:0010032 |
meiotic chromosome condensation(GO:0010032) |
| 0.4 |
2.1 |
GO:0071883 |
activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.3 |
1.0 |
GO:0044413 |
evasion or tolerance of host defenses by virus(GO:0019049) positive regulation of transforming growth factor beta3 production(GO:0032916) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.3 |
1.0 |
GO:0000059 |
protein import into nucleus, docking(GO:0000059) |
| 0.3 |
1.0 |
GO:0009174 |
UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.3 |
1.4 |
GO:0045903 |
positive regulation of translational fidelity(GO:0045903) |
| 0.3 |
2.4 |
GO:0000320 |
re-entry into mitotic cell cycle(GO:0000320) |
| 0.3 |
1.3 |
GO:0072675 |
osteoclast fusion(GO:0072675) |
| 0.3 |
3.3 |
GO:0009396 |
folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.3 |
1.3 |
GO:2000346 |
negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.3 |
1.3 |
GO:0035606 |
peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.3 |
1.0 |
GO:0014028 |
notochord formation(GO:0014028) axial mesoderm morphogenesis(GO:0048319) |
| 0.3 |
1.3 |
GO:0046121 |
deoxyribonucleoside catabolic process(GO:0046121) |
| 0.3 |
2.9 |
GO:0045719 |
negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.3 |
1.0 |
GO:0051096 |
positive regulation of helicase activity(GO:0051096) |
| 0.3 |
1.6 |
GO:0042997 |
negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.3 |
0.3 |
GO:0003162 |
atrioventricular node development(GO:0003162) |
| 0.3 |
1.0 |
GO:0006428 |
isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.3 |
1.2 |
GO:0050862 |
positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.3 |
8.4 |
GO:0034508 |
centromere complex assembly(GO:0034508) |
| 0.3 |
1.5 |
GO:0007223 |
Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.3 |
0.9 |
GO:2001032 |
regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.3 |
1.5 |
GO:0014719 |
skeletal muscle satellite cell activation(GO:0014719) |
| 0.3 |
0.3 |
GO:1903351 |
response to dopamine(GO:1903350) cellular response to dopamine(GO:1903351) |
| 0.3 |
0.9 |
GO:0090526 |
regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.3 |
0.6 |
GO:0010873 |
positive regulation of cholesterol esterification(GO:0010873) |
| 0.3 |
3.3 |
GO:0000463 |
maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.3 |
1.9 |
GO:2000348 |
regulation of CD40 signaling pathway(GO:2000348) |
| 0.3 |
1.7 |
GO:0007253 |
cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.3 |
0.8 |
GO:0060821 |
inactivation of X chromosome by DNA methylation(GO:0060821) |
| 0.3 |
0.8 |
GO:0002572 |
pro-T cell differentiation(GO:0002572) |
| 0.3 |
1.3 |
GO:0034421 |
post-translational protein acetylation(GO:0034421) |
| 0.3 |
0.8 |
GO:0090403 |
regulation of purine nucleotide catabolic process(GO:0033121) negative regulation of purine nucleotide catabolic process(GO:0033122) regulation of fermentation(GO:0043465) oxidative stress-induced premature senescence(GO:0090403) negative regulation of fermentation(GO:1901003) |
| 0.3 |
4.3 |
GO:0015693 |
magnesium ion transport(GO:0015693) |
| 0.3 |
0.8 |
GO:0072718 |
response to cisplatin(GO:0072718) |
| 0.3 |
1.1 |
GO:2000525 |
regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.3 |
1.8 |
GO:0006363 |
termination of RNA polymerase I transcription(GO:0006363) |
| 0.3 |
1.8 |
GO:0042148 |
strand invasion(GO:0042148) |
| 0.3 |
3.9 |
GO:0051984 |
positive regulation of chromosome segregation(GO:0051984) |
| 0.3 |
0.8 |
GO:0051031 |
tRNA transport(GO:0051031) |
| 0.3 |
0.5 |
GO:0010825 |
positive regulation of centrosome duplication(GO:0010825) positive regulation of centrosome cycle(GO:0046607) |
| 0.3 |
1.8 |
GO:0021780 |
oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.2 |
1.2 |
GO:0032298 |
positive regulation of DNA-dependent DNA replication initiation(GO:0032298) |
| 0.2 |
2.0 |
GO:0044351 |
macropinocytosis(GO:0044351) |
| 0.2 |
1.0 |
GO:0006742 |
NADP catabolic process(GO:0006742) pyridine nucleotide catabolic process(GO:0019364) |
| 0.2 |
1.7 |
GO:0007076 |
mitotic chromosome condensation(GO:0007076) |
| 0.2 |
1.2 |
GO:0072429 |
response to cell cycle checkpoint signaling(GO:0072396) response to DNA integrity checkpoint signaling(GO:0072402) response to DNA damage checkpoint signaling(GO:0072423) response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.2 |
2.2 |
GO:0016446 |
somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.2 |
0.2 |
GO:0009414 |
response to water deprivation(GO:0009414) |
| 0.2 |
0.9 |
GO:1990379 |
lipid transport across blood brain barrier(GO:1990379) |
| 0.2 |
0.2 |
GO:2000298 |
regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.2 |
0.7 |
GO:0003360 |
brainstem development(GO:0003360) |
| 0.2 |
0.2 |
GO:0046878 |
positive regulation of saliva secretion(GO:0046878) |
| 0.2 |
1.4 |
GO:0000022 |
mitotic spindle elongation(GO:0000022) mitotic spindle midzone assembly(GO:0051256) |
| 0.2 |
0.9 |
GO:0071475 |
cellular hyperosmotic salinity response(GO:0071475) |
| 0.2 |
0.9 |
GO:0070345 |
negative regulation of fat cell proliferation(GO:0070345) |
| 0.2 |
0.9 |
GO:1902990 |
mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 0.2 |
0.7 |
GO:1900864 |
mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.2 |
0.4 |
GO:0060693 |
regulation of branching involved in salivary gland morphogenesis(GO:0060693) |
| 0.2 |
2.0 |
GO:0071763 |
nuclear membrane organization(GO:0071763) |
| 0.2 |
0.7 |
GO:0042822 |
vitamin B6 metabolic process(GO:0042816) pyridoxal phosphate metabolic process(GO:0042822) pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.2 |
0.7 |
GO:1902915 |
negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.2 |
0.9 |
GO:0051311 |
spindle assembly involved in female meiosis(GO:0007056) meiotic metaphase plate congression(GO:0051311) |
| 0.2 |
0.4 |
GO:0061484 |
hematopoietic stem cell homeostasis(GO:0061484) |
| 0.2 |
1.1 |
GO:0046600 |
negative regulation of centriole replication(GO:0046600) |
| 0.2 |
1.3 |
GO:0002681 |
somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.2 |
0.4 |
GO:1902498 |
regulation of protein autoubiquitination(GO:1902498) |
| 0.2 |
0.9 |
GO:1904749 |
regulation of protein localization to nucleolus(GO:1904749) positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.2 |
0.6 |
GO:0070213 |
protein auto-ADP-ribosylation(GO:0070213) |
| 0.2 |
1.9 |
GO:0050957 |
equilibrioception(GO:0050957) |
| 0.2 |
1.3 |
GO:0090399 |
replicative senescence(GO:0090399) |
| 0.2 |
0.6 |
GO:0097089 |
methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.2 |
2.5 |
GO:0006268 |
DNA unwinding involved in DNA replication(GO:0006268) |
| 0.2 |
0.8 |
GO:0030222 |
eosinophil differentiation(GO:0030222) |
| 0.2 |
0.6 |
GO:0001827 |
inner cell mass cell fate commitment(GO:0001827) |
| 0.2 |
0.8 |
GO:0060032 |
notochord regression(GO:0060032) |
| 0.2 |
0.6 |
GO:0048352 |
neural plate mediolateral regionalization(GO:0021998) mesoderm structural organization(GO:0048338) paraxial mesoderm structural organization(GO:0048352) |
| 0.2 |
0.6 |
GO:1900164 |
nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of transcription from RNA polymerase II promoter involved in determination of left/right symmetry(GO:1900094) nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900164) |
| 0.2 |
2.6 |
GO:0030903 |
notochord development(GO:0030903) |
| 0.2 |
1.8 |
GO:0048733 |
sebaceous gland development(GO:0048733) |
| 0.2 |
2.5 |
GO:0090005 |
negative regulation of establishment of protein localization to plasma membrane(GO:0090005) |
| 0.2 |
1.8 |
GO:0060136 |
embryonic process involved in female pregnancy(GO:0060136) |
| 0.2 |
1.0 |
GO:1904469 |
positive regulation of tumor necrosis factor secretion(GO:1904469) |
| 0.2 |
1.3 |
GO:0007320 |
insemination(GO:0007320) |
| 0.2 |
0.7 |
GO:0018095 |
protein polyglutamylation(GO:0018095) |
| 0.2 |
0.2 |
GO:0031118 |
snoRNA guided rRNA pseudouridine synthesis(GO:0000454) rRNA pseudouridine synthesis(GO:0031118) |
| 0.2 |
2.2 |
GO:0060710 |
chorio-allantoic fusion(GO:0060710) |
| 0.2 |
1.5 |
GO:0009301 |
snRNA transcription(GO:0009301) |
| 0.2 |
0.9 |
GO:0015867 |
ATP transport(GO:0015867) |
| 0.2 |
1.6 |
GO:0035562 |
negative regulation of chromatin binding(GO:0035562) |
| 0.2 |
0.5 |
GO:0002447 |
eosinophil activation involved in immune response(GO:0002278) eosinophil mediated immunity(GO:0002447) eosinophil degranulation(GO:0043308) regulation of eosinophil activation(GO:1902566) |
| 0.2 |
0.7 |
GO:0015793 |
glycerol transport(GO:0015793) |
| 0.2 |
0.5 |
GO:0043696 |
dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.2 |
0.7 |
GO:0046469 |
platelet activating factor biosynthetic process(GO:0006663) platelet activating factor metabolic process(GO:0046469) |
| 0.2 |
0.8 |
GO:0035331 |
negative regulation of hippo signaling(GO:0035331) |
| 0.2 |
0.7 |
GO:0003365 |
establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.2 |
0.7 |
GO:0019287 |
isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.2 |
0.5 |
GO:0097278 |
complement-dependent cytotoxicity(GO:0097278) |
| 0.2 |
1.3 |
GO:0031053 |
primary miRNA processing(GO:0031053) |
| 0.2 |
0.9 |
GO:0009235 |
cobalamin metabolic process(GO:0009235) |
| 0.2 |
0.5 |
GO:0021577 |
hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.2 |
0.9 |
GO:0042256 |
mature ribosome assembly(GO:0042256) |
| 0.2 |
4.3 |
GO:0009409 |
response to cold(GO:0009409) |
| 0.2 |
0.8 |
GO:0090666 |
scaRNA localization to Cajal body(GO:0090666) |
| 0.2 |
0.9 |
GO:0010804 |
negative regulation of tumor necrosis factor-mediated signaling pathway(GO:0010804) |
| 0.2 |
0.9 |
GO:0085020 |
protein K6-linked ubiquitination(GO:0085020) |
| 0.1 |
0.9 |
GO:0071294 |
cellular response to zinc ion(GO:0071294) |
| 0.1 |
2.4 |
GO:0006270 |
DNA replication initiation(GO:0006270) |
| 0.1 |
0.4 |
GO:1902445 |
negative regulation of oxidative phosphorylation(GO:0090324) regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.1 |
0.4 |
GO:0044339 |
canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.1 |
2.2 |
GO:0032508 |
DNA duplex unwinding(GO:0032508) |
| 0.1 |
1.6 |
GO:0038092 |
nodal signaling pathway(GO:0038092) |
| 0.1 |
1.0 |
GO:0016584 |
nucleosome positioning(GO:0016584) |
| 0.1 |
0.6 |
GO:0097211 |
response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.1 |
0.4 |
GO:1903546 |
protein localization to photoreceptor outer segment(GO:1903546) |
| 0.1 |
0.4 |
GO:2000686 |
regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.1 |
0.6 |
GO:0034239 |
regulation of macrophage fusion(GO:0034239) |
| 0.1 |
1.4 |
GO:0036123 |
histone H3-K9 dimethylation(GO:0036123) |
| 0.1 |
0.4 |
GO:0035574 |
histone H4-K20 demethylation(GO:0035574) |
| 0.1 |
3.4 |
GO:0006607 |
NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 |
0.7 |
GO:0006297 |
nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.1 |
2.3 |
GO:0007530 |
sex determination(GO:0007530) |
| 0.1 |
0.4 |
GO:1903847 |
regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 0.1 |
0.6 |
GO:0006447 |
regulation of translational initiation by iron(GO:0006447) positive regulation of translational initiation by iron(GO:0045994) |
| 0.1 |
1.4 |
GO:1903333 |
negative regulation of protein folding(GO:1903333) |
| 0.1 |
0.3 |
GO:0050883 |
musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.1 |
0.4 |
GO:0046077 |
dUDP biosynthetic process(GO:0006227) pyrimidine nucleoside diphosphate biosynthetic process(GO:0009139) pyrimidine deoxyribonucleoside diphosphate metabolic process(GO:0009196) pyrimidine deoxyribonucleoside diphosphate biosynthetic process(GO:0009197) dUDP metabolic process(GO:0046077) |
| 0.1 |
0.8 |
GO:0035428 |
hexose transmembrane transport(GO:0035428) |
| 0.1 |
1.4 |
GO:0036297 |
interstrand cross-link repair(GO:0036297) |
| 0.1 |
1.1 |
GO:0010815 |
bradykinin catabolic process(GO:0010815) |
| 0.1 |
3.4 |
GO:0008299 |
isoprenoid biosynthetic process(GO:0008299) |
| 0.1 |
0.5 |
GO:0035795 |
negative regulation of mitochondrial membrane permeability(GO:0035795) |
| 0.1 |
2.6 |
GO:0071539 |
protein localization to centrosome(GO:0071539) |
| 0.1 |
0.5 |
GO:0071033 |
nuclear retention of pre-mRNA at the site of transcription(GO:0071033) CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.1 |
0.9 |
GO:0033184 |
positive regulation of histone ubiquitination(GO:0033184) |
| 0.1 |
0.3 |
GO:0034196 |
acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.1 |
2.1 |
GO:0034453 |
microtubule anchoring(GO:0034453) |
| 0.1 |
0.9 |
GO:0000414 |
regulation of histone H3-K36 methylation(GO:0000414) |
| 0.1 |
0.9 |
GO:0007296 |
vitellogenesis(GO:0007296) |
| 0.1 |
0.9 |
GO:0032790 |
ribosome disassembly(GO:0032790) |
| 0.1 |
0.8 |
GO:0060330 |
regulation of response to interferon-gamma(GO:0060330) |
| 0.1 |
0.8 |
GO:0006177 |
GMP biosynthetic process(GO:0006177) |
| 0.1 |
0.9 |
GO:0043569 |
negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.1 |
0.6 |
GO:0090244 |
Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.1 |
1.8 |
GO:0048642 |
negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.1 |
0.5 |
GO:0070940 |
dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.1 |
0.4 |
GO:1902626 |
assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.1 |
0.4 |
GO:0060010 |
Sertoli cell fate commitment(GO:0060010) |
| 0.1 |
0.4 |
GO:0015882 |
L-ascorbic acid transport(GO:0015882) dehydroascorbic acid transport(GO:0070837) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.1 |
1.1 |
GO:0090435 |
protein localization to nuclear envelope(GO:0090435) |
| 0.1 |
0.4 |
GO:0046032 |
ADP catabolic process(GO:0046032) |
| 0.1 |
0.7 |
GO:0098700 |
aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.1 |
0.6 |
GO:0070562 |
regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.1 |
0.5 |
GO:0019614 |
catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) |
| 0.1 |
0.2 |
GO:0039530 |
MDA-5 signaling pathway(GO:0039530) |
| 0.1 |
0.7 |
GO:0044829 |
positive regulation by host of viral genome replication(GO:0044829) |
| 0.1 |
0.4 |
GO:0060060 |
post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.1 |
0.4 |
GO:0048496 |
maintenance of organ identity(GO:0048496) |
| 0.1 |
1.4 |
GO:0044458 |
motile cilium assembly(GO:0044458) |
| 0.1 |
1.8 |
GO:0035414 |
negative regulation of catenin import into nucleus(GO:0035414) |
| 0.1 |
1.6 |
GO:0051044 |
positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 |
0.8 |
GO:0051639 |
actin filament network formation(GO:0051639) |
| 0.1 |
0.5 |
GO:0001306 |
age-dependent response to oxidative stress(GO:0001306) age-dependent general metabolic decline(GO:0007571) |
| 0.1 |
0.3 |
GO:1903750 |
regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) |
| 0.1 |
1.3 |
GO:2000381 |
negative regulation of mesoderm development(GO:2000381) |
| 0.1 |
0.3 |
GO:0006592 |
ornithine biosynthetic process(GO:0006592) |
| 0.1 |
0.6 |
GO:0006621 |
protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.1 |
0.2 |
GO:0003100 |
regulation of systemic arterial blood pressure by endothelin(GO:0003100) |
| 0.1 |
1.5 |
GO:0006298 |
mismatch repair(GO:0006298) |
| 0.1 |
0.3 |
GO:0070535 |
histone H2A K63-linked ubiquitination(GO:0070535) |
| 0.1 |
1.1 |
GO:0014807 |
regulation of somitogenesis(GO:0014807) |
| 0.1 |
0.5 |
GO:1900246 |
positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.1 |
0.5 |
GO:0007144 |
female meiosis I(GO:0007144) |
| 0.1 |
0.7 |
GO:0060028 |
convergent extension involved in axis elongation(GO:0060028) |
| 0.1 |
2.1 |
GO:0061157 |
mRNA destabilization(GO:0061157) |
| 0.1 |
1.0 |
GO:0045086 |
positive regulation of interleukin-2 biosynthetic process(GO:0045086) |
| 0.1 |
0.7 |
GO:1903755 |
regulation of SUMO transferase activity(GO:1903182) positive regulation of SUMO transferase activity(GO:1903755) |
| 0.1 |
1.0 |
GO:0010803 |
regulation of tumor necrosis factor-mediated signaling pathway(GO:0010803) |
| 0.1 |
0.3 |
GO:0045925 |
positive regulation of female receptivity(GO:0045925) |
| 0.1 |
0.9 |
GO:0006271 |
DNA strand elongation involved in DNA replication(GO:0006271) |
| 0.1 |
0.2 |
GO:1901165 |
positive regulation of trophoblast cell migration(GO:1901165) |
| 0.1 |
0.3 |
GO:0000087 |
mitotic M phase(GO:0000087) |
| 0.1 |
0.3 |
GO:0034729 |
histone H3-K79 methylation(GO:0034729) |
| 0.1 |
0.5 |
GO:0030091 |
protein repair(GO:0030091) |
| 0.1 |
0.6 |
GO:0046909 |
intermembrane transport(GO:0046909) |
| 0.1 |
0.6 |
GO:0007016 |
cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.1 |
0.3 |
GO:1902396 |
protein localization to bicellular tight junction(GO:1902396) |
| 0.1 |
0.4 |
GO:0008616 |
queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.1 |
0.3 |
GO:0033860 |
regulation of NAD(P)H oxidase activity(GO:0033860) |
| 0.1 |
1.6 |
GO:0042407 |
cristae formation(GO:0042407) |
| 0.1 |
0.7 |
GO:0070458 |
detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 0.1 |
1.7 |
GO:0001522 |
pseudouridine synthesis(GO:0001522) |
| 0.1 |
0.4 |
GO:0033580 |
protein glycosylation at cell surface(GO:0033575) protein galactosylation at cell surface(GO:0033580) protein galactosylation(GO:0042125) |
| 0.1 |
0.8 |
GO:1904816 |
positive regulation of protein localization to chromosome, telomeric region(GO:1904816) |
| 0.1 |
1.7 |
GO:0031442 |
positive regulation of mRNA 3'-end processing(GO:0031442) |
| 0.1 |
1.1 |
GO:0060539 |
diaphragm development(GO:0060539) |
| 0.1 |
0.9 |
GO:0061179 |
negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.1 |
0.3 |
GO:0042637 |
catagen(GO:0042637) |
| 0.1 |
0.8 |
GO:0031936 |
negative regulation of chromatin silencing(GO:0031936) |
| 0.1 |
0.7 |
GO:0016127 |
cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.1 |
0.4 |
GO:0032781 |
positive regulation of ATPase activity(GO:0032781) |
| 0.1 |
2.4 |
GO:0000460 |
maturation of 5.8S rRNA(GO:0000460) |
| 0.1 |
0.7 |
GO:1902231 |
positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.1 |
0.7 |
GO:0055064 |
carbon dioxide transport(GO:0015670) chloride ion homeostasis(GO:0055064) |
| 0.1 |
0.3 |
GO:0006361 |
transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.1 |
0.7 |
GO:0090074 |
negative regulation of protein homodimerization activity(GO:0090074) |
| 0.1 |
0.6 |
GO:0031440 |
regulation of mRNA 3'-end processing(GO:0031440) |
| 0.1 |
0.4 |
GO:0042276 |
error-prone translesion synthesis(GO:0042276) |
| 0.1 |
0.5 |
GO:1902965 |
regulation of protein localization to early endosome(GO:1902965) positive regulation of protein localization to early endosome(GO:1902966) |
| 0.1 |
0.4 |
GO:0033629 |
negative regulation of cell adhesion mediated by integrin(GO:0033629) |
| 0.1 |
0.4 |
GO:0090370 |
negative regulation of cholesterol efflux(GO:0090370) |
| 0.1 |
0.3 |
GO:0031590 |
wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.1 |
0.3 |
GO:0006776 |
vitamin A metabolic process(GO:0006776) |
| 0.1 |
0.2 |
GO:0046501 |
protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.1 |
0.1 |
GO:0034140 |
negative regulation of toll-like receptor 3 signaling pathway(GO:0034140) |
| 0.1 |
0.4 |
GO:0090110 |
cargo loading into COPII-coated vesicle(GO:0090110) regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.1 |
0.7 |
GO:0006398 |
mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.1 |
0.5 |
GO:0090073 |
positive regulation of protein homodimerization activity(GO:0090073) |
| 0.1 |
0.3 |
GO:0010878 |
cholesterol storage(GO:0010878) |
| 0.1 |
0.6 |
GO:0001887 |
selenium compound metabolic process(GO:0001887) |
| 0.1 |
0.5 |
GO:2000210 |
positive regulation of anoikis(GO:2000210) |
| 0.1 |
0.6 |
GO:1900262 |
regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.1 |
1.4 |
GO:1902043 |
positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.1 |
0.6 |
GO:0035735 |
intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.1 |
0.5 |
GO:0061052 |
negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 0.1 |
0.6 |
GO:0007000 |
nucleolus organization(GO:0007000) |
| 0.1 |
0.6 |
GO:0007195 |
adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.1 |
0.2 |
GO:0002035 |
brain renin-angiotensin system(GO:0002035) |
| 0.1 |
0.4 |
GO:0001867 |
complement activation, lectin pathway(GO:0001867) |
| 0.1 |
0.3 |
GO:0044861 |
protein transport into plasma membrane raft(GO:0044861) |
| 0.1 |
0.4 |
GO:0090502 |
RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.1 |
0.2 |
GO:0006627 |
protein processing involved in protein targeting to mitochondrion(GO:0006627) mitochondrial protein processing(GO:0034982) |
| 0.1 |
0.5 |
GO:0008611 |
ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) cellular lipid biosynthetic process(GO:0097384) ether biosynthetic process(GO:1901503) |
| 0.1 |
0.9 |
GO:0006000 |
fructose metabolic process(GO:0006000) |
| 0.1 |
1.3 |
GO:0006907 |
pinocytosis(GO:0006907) |
| 0.1 |
1.3 |
GO:0007080 |
mitotic metaphase plate congression(GO:0007080) |
| 0.1 |
0.8 |
GO:0043153 |
entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.1 |
0.6 |
GO:0021796 |
cerebral cortex regionalization(GO:0021796) |
| 0.1 |
1.1 |
GO:0090261 |
positive regulation of inclusion body assembly(GO:0090261) |
| 0.1 |
1.1 |
GO:0000188 |
inactivation of MAPK activity(GO:0000188) |
| 0.1 |
1.8 |
GO:0044819 |
mitotic G1/S transition checkpoint(GO:0044819) |
| 0.1 |
0.9 |
GO:0046697 |
decidualization(GO:0046697) |
| 0.1 |
0.2 |
GO:0009051 |
pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.1 |
0.2 |
GO:2000834 |
luteinizing hormone secretion(GO:0032275) androgen secretion(GO:0035935) testosterone secretion(GO:0035936) regulation of androgen secretion(GO:2000834) positive regulation of androgen secretion(GO:2000836) regulation of testosterone secretion(GO:2000843) positive regulation of testosterone secretion(GO:2000845) |
| 0.1 |
0.7 |
GO:0030035 |
microspike assembly(GO:0030035) |
| 0.1 |
0.4 |
GO:0071622 |
regulation of granulocyte chemotaxis(GO:0071622) |
| 0.1 |
0.4 |
GO:0051310 |
metaphase plate congression(GO:0051310) |
| 0.1 |
1.0 |
GO:0006924 |
activation-induced cell death of T cells(GO:0006924) |
| 0.1 |
0.4 |
GO:0042780 |
tRNA 3'-end processing(GO:0042780) |
| 0.1 |
0.7 |
GO:0030261 |
chromosome condensation(GO:0030261) |
| 0.1 |
0.4 |
GO:0001778 |
plasma membrane repair(GO:0001778) |
| 0.1 |
0.5 |
GO:0006384 |
transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.1 |
0.6 |
GO:0009048 |
dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.1 |
1.0 |
GO:0030150 |
protein import into mitochondrial matrix(GO:0030150) |
| 0.1 |
0.1 |
GO:0072106 |
regulation of ureteric bud formation(GO:0072106) positive regulation of ureteric bud formation(GO:0072107) |
| 0.1 |
0.6 |
GO:0042730 |
fibrinolysis(GO:0042730) |
| 0.1 |
0.1 |
GO:1904211 |
membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.1 |
0.9 |
GO:0048672 |
positive regulation of collateral sprouting(GO:0048672) |
| 0.1 |
0.2 |
GO:0021986 |
epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.1 |
1.1 |
GO:0050892 |
intestinal absorption(GO:0050892) |
| 0.1 |
0.1 |
GO:0003406 |
retinal pigment epithelium development(GO:0003406) |
| 0.1 |
1.6 |
GO:0033209 |
tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.1 |
0.2 |
GO:1904729 |
regulation of intestinal cholesterol absorption(GO:0030300) regulation of intestinal absorption(GO:1904478) regulation of intestinal lipid absorption(GO:1904729) |
| 0.1 |
0.3 |
GO:0000289 |
nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.1 |
1.2 |
GO:0048535 |
lymph node development(GO:0048535) |
| 0.1 |
0.6 |
GO:0048739 |
cardiac muscle fiber development(GO:0048739) |
| 0.1 |
4.1 |
GO:0051225 |
spindle assembly(GO:0051225) |
| 0.1 |
0.6 |
GO:0032048 |
cardiolipin metabolic process(GO:0032048) |
| 0.1 |
0.3 |
GO:0030866 |
cortical actin cytoskeleton organization(GO:0030866) |
| 0.1 |
0.8 |
GO:0000244 |
spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.1 |
0.3 |
GO:2000258 |
negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.1 |
2.5 |
GO:0007026 |
negative regulation of microtubule depolymerization(GO:0007026) |
| 0.1 |
0.1 |
GO:2000286 |
receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.1 |
0.4 |
GO:0008635 |
activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.1 |
0.7 |
GO:0042711 |
maternal behavior(GO:0042711) |
| 0.1 |
1.1 |
GO:0033687 |
osteoblast proliferation(GO:0033687) |
| 0.1 |
0.3 |
GO:0032911 |
negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.1 |
0.5 |
GO:0035845 |
photoreceptor cell outer segment organization(GO:0035845) |
| 0.1 |
0.4 |
GO:1901724 |
positive regulation of cell proliferation involved in kidney development(GO:1901724) |
| 0.1 |
0.8 |
GO:0042219 |
cellular modified amino acid catabolic process(GO:0042219) |
| 0.1 |
0.2 |
GO:0070889 |
platelet alpha granule organization(GO:0070889) |
| 0.1 |
0.5 |
GO:0070935 |
3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.1 |
0.3 |
GO:0045723 |
positive regulation of fatty acid biosynthetic process(GO:0045723) |
| 0.1 |
1.0 |
GO:0070050 |
neuron cellular homeostasis(GO:0070050) |
| 0.1 |
0.6 |
GO:0035721 |
intraciliary retrograde transport(GO:0035721) |
| 0.1 |
0.2 |
GO:0006436 |
tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.1 |
0.5 |
GO:2000467 |
positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.1 |
0.2 |
GO:0019085 |
early viral transcription(GO:0019085) |
| 0.1 |
0.7 |
GO:0051292 |
nuclear pore complex assembly(GO:0051292) |
| 0.1 |
1.1 |
GO:0097150 |
neuronal stem cell population maintenance(GO:0097150) |
| 0.1 |
0.4 |
GO:0045948 |
positive regulation of translational initiation(GO:0045948) |
| 0.1 |
0.6 |
GO:0045947 |
negative regulation of translational initiation(GO:0045947) |
| 0.1 |
0.1 |
GO:0006295 |
nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.1 |
0.1 |
GO:0071594 |
T cell differentiation in thymus(GO:0033077) thymocyte aggregation(GO:0071594) |
| 0.1 |
0.8 |
GO:0046686 |
response to cadmium ion(GO:0046686) |
| 0.1 |
0.3 |
GO:0051697 |
protein delipidation(GO:0051697) |
| 0.1 |
0.2 |
GO:0003349 |
epicardium-derived cardiac endothelial cell differentiation(GO:0003349) |
| 0.1 |
0.2 |
GO:0016095 |
polyprenol catabolic process(GO:0016095) |
| 0.1 |
0.6 |
GO:0042347 |
negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.1 |
0.4 |
GO:0035948 |
positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) regulation of cellular ketone metabolic process by positive regulation of transcription from RNA polymerase II promoter(GO:0072366) |
| 0.1 |
0.9 |
GO:0019430 |
removal of superoxide radicals(GO:0019430) |
| 0.1 |
0.2 |
GO:0001542 |
ovulation from ovarian follicle(GO:0001542) |
| 0.1 |
0.8 |
GO:0009954 |
proximal/distal pattern formation(GO:0009954) |
| 0.1 |
0.2 |
GO:0007084 |
mitotic nuclear envelope reassembly(GO:0007084) |
| 0.1 |
0.2 |
GO:0002834 |
immune response to tumor cell(GO:0002418) regulation of response to tumor cell(GO:0002834) regulation of immune response to tumor cell(GO:0002837) |
| 0.1 |
0.3 |
GO:0090164 |
asymmetric Golgi ribbon formation(GO:0090164) |
| 0.1 |
3.4 |
GO:0002181 |
cytoplasmic translation(GO:0002181) |
| 0.1 |
1.3 |
GO:0006446 |
regulation of translational initiation(GO:0006446) |
| 0.1 |
0.5 |
GO:0021521 |
ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.1 |
0.7 |
GO:0019368 |
fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 |
0.1 |
GO:0097155 |
fasciculation of sensory neuron axon(GO:0097155) |
| 0.1 |
0.8 |
GO:0006388 |
tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.1 |
0.9 |
GO:0000028 |
ribosomal small subunit assembly(GO:0000028) |
| 0.1 |
0.6 |
GO:0010225 |
response to UV-C(GO:0010225) |
| 0.1 |
0.2 |
GO:2000773 |
negative regulation of cellular senescence(GO:2000773) |
| 0.1 |
0.2 |
GO:0030579 |
ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.1 |
0.6 |
GO:0071157 |
negative regulation of cell cycle arrest(GO:0071157) |
| 0.1 |
0.8 |
GO:0035493 |
SNARE complex assembly(GO:0035493) |
| 0.1 |
0.2 |
GO:0032382 |
positive regulation of intracellular lipid transport(GO:0032379) positive regulation of intracellular sterol transport(GO:0032382) positive regulation of intracellular cholesterol transport(GO:0032385) lipid hydroperoxide transport(GO:1901373) positive regulation of cholesterol import(GO:1904109) positive regulation of sterol import(GO:2000911) |
| 0.1 |
0.2 |
GO:0009644 |
response to high light intensity(GO:0009644) |
| 0.1 |
0.8 |
GO:0070816 |
phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.1 |
0.7 |
GO:2000480 |
negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 |
0.2 |
GO:0045204 |
MAPK export from nucleus(GO:0045204) |
| 0.1 |
0.2 |
GO:0006421 |
asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.1 |
0.7 |
GO:2000060 |
positive regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000060) |
| 0.1 |
0.2 |
GO:0018094 |
protein polyglycylation(GO:0018094) |
| 0.1 |
2.9 |
GO:0010389 |
regulation of G2/M transition of mitotic cell cycle(GO:0010389) |
| 0.1 |
2.4 |
GO:0010501 |
RNA secondary structure unwinding(GO:0010501) |
| 0.1 |
1.2 |
GO:0000413 |
protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 |
0.2 |
GO:0033683 |
nucleotide-excision repair, DNA incision(GO:0033683) |
| 0.1 |
0.1 |
GO:0010826 |
negative regulation of centrosome duplication(GO:0010826) negative regulation of centrosome cycle(GO:0046606) |
| 0.1 |
0.3 |
GO:0034587 |
piRNA metabolic process(GO:0034587) |
| 0.1 |
0.1 |
GO:0000722 |
telomere maintenance via recombination(GO:0000722) |
| 0.1 |
0.7 |
GO:0018126 |
protein hydroxylation(GO:0018126) |
| 0.1 |
1.2 |
GO:0042462 |
eye photoreceptor cell development(GO:0042462) |
| 0.1 |
0.4 |
GO:0061525 |
hindgut development(GO:0061525) |
| 0.1 |
1.7 |
GO:0000381 |
regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 |
0.2 |
GO:0007406 |
negative regulation of neuroblast proliferation(GO:0007406) |
| 0.1 |
0.4 |
GO:0051571 |
positive regulation of histone H3-K4 methylation(GO:0051571) |
| 0.1 |
0.5 |
GO:0061302 |
smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.1 |
1.9 |
GO:1902476 |
chloride transmembrane transport(GO:1902476) |
| 0.1 |
0.8 |
GO:0006465 |
signal peptide processing(GO:0006465) |
| 0.1 |
0.6 |
GO:0015825 |
L-serine transport(GO:0015825) |
| 0.0 |
0.9 |
GO:0046329 |
negative regulation of JNK cascade(GO:0046329) |
| 0.0 |
0.2 |
GO:0051490 |
negative regulation of filopodium assembly(GO:0051490) |
| 0.0 |
0.2 |
GO:0046294 |
formaldehyde catabolic process(GO:0046294) |
| 0.0 |
0.4 |
GO:0007175 |
negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 |
0.2 |
GO:0021764 |
amygdala development(GO:0021764) |
| 0.0 |
0.4 |
GO:0080009 |
mRNA methylation(GO:0080009) |
| 0.0 |
0.5 |
GO:0008105 |
asymmetric protein localization(GO:0008105) |
| 0.0 |
1.2 |
GO:0000462 |
maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 |
1.1 |
GO:0030488 |
tRNA methylation(GO:0030488) |
| 0.0 |
0.3 |
GO:0043619 |
regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.0 |
0.7 |
GO:0051123 |
RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) DNA-templated transcriptional preinitiation complex assembly(GO:0070897) |
| 0.0 |
0.2 |
GO:0060040 |
retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 |
0.4 |
GO:0018026 |
peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 |
0.3 |
GO:0051694 |
pointed-end actin filament capping(GO:0051694) |
| 0.0 |
0.4 |
GO:0010571 |
positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.0 |
0.4 |
GO:0042095 |
interferon-gamma biosynthetic process(GO:0042095) |
| 0.0 |
0.3 |
GO:0006449 |
regulation of translational termination(GO:0006449) translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.0 |
0.1 |
GO:0032053 |
ciliary basal body organization(GO:0032053) |
| 0.0 |
0.4 |
GO:0001675 |
acrosome assembly(GO:0001675) |
| 0.0 |
1.5 |
GO:0021591 |
ventricular system development(GO:0021591) |
| 0.0 |
0.4 |
GO:0007250 |
activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 |
0.2 |
GO:0070966 |
nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 |
0.4 |
GO:0036260 |
7-methylguanosine RNA capping(GO:0009452) RNA capping(GO:0036260) |
| 0.0 |
0.7 |
GO:0060009 |
Sertoli cell development(GO:0060009) |
| 0.0 |
0.6 |
GO:0003334 |
keratinocyte development(GO:0003334) |
| 0.0 |
0.3 |
GO:1903352 |
ornithine transport(GO:0015822) L-ornithine transmembrane transport(GO:1903352) |
| 0.0 |
0.4 |
GO:0030049 |
muscle filament sliding(GO:0030049) |
| 0.0 |
0.2 |
GO:0071557 |
histone H3-K27 demethylation(GO:0071557) |
| 0.0 |
1.0 |
GO:0018345 |
protein palmitoylation(GO:0018345) |
| 0.0 |
0.2 |
GO:0040032 |
post-embryonic body morphogenesis(GO:0040032) |
| 0.0 |
0.2 |
GO:0060575 |
intestinal epithelial cell differentiation(GO:0060575) |
| 0.0 |
0.5 |
GO:2000009 |
negative regulation of protein localization to cell surface(GO:2000009) |
| 0.0 |
0.5 |
GO:0009083 |
branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 |
0.2 |
GO:1902224 |
cellular ketone body metabolic process(GO:0046950) ketone body metabolic process(GO:1902224) |
| 0.0 |
0.2 |
GO:0001980 |
regulation of systemic arterial blood pressure by ischemic conditions(GO:0001980) |
| 0.0 |
0.3 |
GO:0032927 |
positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 |
0.1 |
GO:0009138 |
pyrimidine nucleoside diphosphate metabolic process(GO:0009138) |
| 0.0 |
0.3 |
GO:0015074 |
DNA integration(GO:0015074) |
| 0.0 |
1.3 |
GO:0005978 |
glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 |
0.1 |
GO:0060008 |
Sertoli cell differentiation(GO:0060008) |
| 0.0 |
0.1 |
GO:2001013 |
epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 |
0.6 |
GO:0006910 |
phagocytosis, recognition(GO:0006910) |
| 0.0 |
0.1 |
GO:0051088 |
PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.0 |
0.3 |
GO:0021942 |
radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.0 |
1.1 |
GO:0051930 |
regulation of sensory perception of pain(GO:0051930) regulation of sensory perception(GO:0051931) |
| 0.0 |
0.7 |
GO:0045909 |
positive regulation of vasodilation(GO:0045909) |
| 0.0 |
0.4 |
GO:0061684 |
chaperone-mediated autophagy(GO:0061684) |
| 0.0 |
0.1 |
GO:0002901 |
mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.0 |
0.5 |
GO:0042517 |
positive regulation of tyrosine phosphorylation of Stat3 protein(GO:0042517) |
| 0.0 |
0.3 |
GO:0034773 |
histone H4-K20 trimethylation(GO:0034773) |
| 0.0 |
0.2 |
GO:0009650 |
UV protection(GO:0009650) |
| 0.0 |
0.1 |
GO:1905049 |
negative regulation of metalloendopeptidase activity(GO:1904684) negative regulation of metallopeptidase activity(GO:1905049) |
| 0.0 |
0.2 |
GO:0031468 |
nuclear envelope reassembly(GO:0031468) |
| 0.0 |
0.1 |
GO:1990173 |
protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) protein localization to nucleoplasm(GO:1990173) |
| 0.0 |
0.4 |
GO:0048280 |
vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 |
0.2 |
GO:0032968 |
positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 |
0.2 |
GO:0070365 |
hepatocyte differentiation(GO:0070365) |
| 0.0 |
0.2 |
GO:0007252 |
I-kappaB phosphorylation(GO:0007252) |
| 0.0 |
0.8 |
GO:0000387 |
spliceosomal snRNP assembly(GO:0000387) |
| 0.0 |
0.4 |
GO:0043486 |
histone exchange(GO:0043486) |
| 0.0 |
0.2 |
GO:0031584 |
activation of phospholipase D activity(GO:0031584) |
| 0.0 |
0.1 |
GO:0060021 |
palate development(GO:0060021) |
| 0.0 |
0.3 |
GO:0051131 |
chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 |
0.7 |
GO:0033627 |
cell adhesion mediated by integrin(GO:0033627) |
| 0.0 |
0.2 |
GO:1902861 |
copper ion import into cell(GO:1902861) |
| 0.0 |
0.2 |
GO:0045654 |
positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.0 |
0.5 |
GO:0006801 |
superoxide metabolic process(GO:0006801) |
| 0.0 |
1.1 |
GO:0061077 |
chaperone-mediated protein folding(GO:0061077) |
| 0.0 |
0.4 |
GO:0010862 |
positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 |
0.6 |
GO:0043968 |
histone H2A acetylation(GO:0043968) |
| 0.0 |
0.7 |
GO:0006284 |
base-excision repair(GO:0006284) |
| 0.0 |
1.1 |
GO:0045600 |
positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 |
0.3 |
GO:0035520 |
monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 |
0.1 |
GO:0090086 |
negative regulation of protein deubiquitination(GO:0090086) |
| 0.0 |
0.2 |
GO:0045713 |
low-density lipoprotein particle receptor biosynthetic process(GO:0045713) regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045714) |
| 0.0 |
0.1 |
GO:1902047 |
polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) |
| 0.0 |
0.1 |
GO:0019065 |
receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.0 |
0.2 |
GO:0003215 |
cardiac right ventricle morphogenesis(GO:0003215) |
| 0.0 |
0.1 |
GO:0006824 |
cobalt ion transport(GO:0006824) |
| 0.0 |
0.3 |
GO:0006379 |
mRNA cleavage(GO:0006379) |
| 0.0 |
0.3 |
GO:0010640 |
regulation of platelet-derived growth factor receptor signaling pathway(GO:0010640) |
| 0.0 |
0.1 |
GO:1903225 |
regulation of endodermal cell differentiation(GO:1903224) negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 |
0.2 |
GO:0006105 |
succinate metabolic process(GO:0006105) |
| 0.0 |
0.1 |
GO:0002934 |
desmosome organization(GO:0002934) |
| 0.0 |
0.1 |
GO:0006983 |
ER overload response(GO:0006983) |
| 0.0 |
0.2 |
GO:0000103 |
sulfate assimilation(GO:0000103) |
| 0.0 |
0.1 |
GO:0002315 |
marginal zone B cell differentiation(GO:0002315) |
| 0.0 |
0.2 |
GO:0006104 |
succinyl-CoA metabolic process(GO:0006104) |
| 0.0 |
0.5 |
GO:0045843 |
negative regulation of striated muscle tissue development(GO:0045843) |
| 0.0 |
0.1 |
GO:0050667 |
homocysteine metabolic process(GO:0050667) |
| 0.0 |
0.8 |
GO:0034644 |
cellular response to UV(GO:0034644) |
| 0.0 |
0.7 |
GO:0032835 |
glomerulus development(GO:0032835) |
| 0.0 |
0.1 |
GO:1903300 |
negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 |
0.2 |
GO:0003341 |
cilium movement(GO:0003341) |
| 0.0 |
0.9 |
GO:0006094 |
gluconeogenesis(GO:0006094) |
| 0.0 |
0.1 |
GO:0034143 |
regulation of toll-like receptor 4 signaling pathway(GO:0034143) |
| 0.0 |
0.2 |
GO:0060416 |
response to growth hormone(GO:0060416) |
| 0.0 |
0.8 |
GO:0071456 |
cellular response to hypoxia(GO:0071456) |
| 0.0 |
0.1 |
GO:0014886 |
transition between slow and fast fiber(GO:0014886) |
| 0.0 |
0.1 |
GO:0018065 |
protein-cofactor linkage(GO:0018065) |
| 0.0 |
0.1 |
GO:0090195 |
chemokine secretion(GO:0090195) regulation of chemokine secretion(GO:0090196) positive regulation of chemokine secretion(GO:0090197) |
| 0.0 |
0.8 |
GO:0061003 |
positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.0 |
0.3 |
GO:0009299 |
mRNA transcription(GO:0009299) |
| 0.0 |
0.2 |
GO:0043278 |
response to isoquinoline alkaloid(GO:0014072) response to morphine(GO:0043278) |
| 0.0 |
0.5 |
GO:0043488 |
regulation of mRNA stability(GO:0043488) |
| 0.0 |
0.1 |
GO:0002036 |
regulation of L-glutamate transport(GO:0002036) |
| 0.0 |
0.0 |
GO:0052428 |
modulation of molecular function in other organism(GO:0044359) negative regulation of molecular function in other organism(GO:0044362) negative regulation of molecular function in other organism involved in symbiotic interaction(GO:0052204) modulation of molecular function in other organism involved in symbiotic interaction(GO:0052205) negative regulation by host of symbiont molecular function(GO:0052405) modification by host of symbiont molecular function(GO:0052428) |
| 0.0 |
0.0 |
GO:0006999 |
nuclear pore organization(GO:0006999) |
| 0.0 |
0.6 |
GO:0042633 |
molting cycle(GO:0042303) hair cycle(GO:0042633) |
| 0.0 |
0.3 |
GO:0070979 |
protein K11-linked ubiquitination(GO:0070979) |
| 0.0 |
0.1 |
GO:0015788 |
UDP-N-acetylglucosamine transport(GO:0015788) |
| 0.0 |
0.2 |
GO:0060394 |
negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 |
0.2 |
GO:0018206 |
peptidyl-methionine modification(GO:0018206) |
| 0.0 |
0.6 |
GO:0007608 |
sensory perception of smell(GO:0007608) |
| 0.0 |
0.6 |
GO:2001222 |
regulation of neuron migration(GO:2001222) |
| 0.0 |
0.2 |
GO:0001672 |
regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 |
0.3 |
GO:0035873 |
lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.0 |
0.2 |
GO:0035058 |
nonmotile primary cilium assembly(GO:0035058) |
| 0.0 |
0.2 |
GO:0046085 |
adenosine metabolic process(GO:0046085) |
| 0.0 |
0.1 |
GO:0050915 |
sensory perception of sour taste(GO:0050915) |
| 0.0 |
0.2 |
GO:0002089 |
lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 |
0.2 |
GO:0035871 |
protein K11-linked deubiquitination(GO:0035871) |
| 0.0 |
0.4 |
GO:0001947 |
heart looping(GO:0001947) |
| 0.0 |
0.2 |
GO:0002385 |
organ or tissue specific immune response(GO:0002251) mucosal immune response(GO:0002385) |
| 0.0 |
0.3 |
GO:0070884 |
regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.0 |
0.3 |
GO:0071425 |
hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 |
0.1 |
GO:0031670 |
cellular response to nutrient(GO:0031670) |
| 0.0 |
0.1 |
GO:0016338 |
calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 |
0.1 |
GO:0035518 |
histone H2A monoubiquitination(GO:0035518) |
| 0.0 |
0.8 |
GO:0045454 |
cell redox homeostasis(GO:0045454) |
| 0.0 |
0.1 |
GO:0007221 |
positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 |
0.3 |
GO:0016226 |
iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 |
0.4 |
GO:0010591 |
regulation of lamellipodium assembly(GO:0010591) |
| 0.0 |
0.1 |
GO:0060837 |
blood vessel endothelial cell differentiation(GO:0060837) |
| 0.0 |
0.2 |
GO:0030317 |
sperm motility(GO:0030317) |
| 0.0 |
0.2 |
GO:0007599 |
blood coagulation(GO:0007596) hemostasis(GO:0007599) |
| 0.0 |
0.1 |
GO:0042732 |
D-xylose metabolic process(GO:0042732) |
| 0.0 |
0.5 |
GO:0048384 |
retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 |
0.5 |
GO:0045682 |
regulation of epidermis development(GO:0045682) |
| 0.0 |
0.1 |
GO:1901020 |
negative regulation of calcium ion transmembrane transporter activity(GO:1901020) negative regulation of calcium ion transmembrane transport(GO:1903170) |
| 0.0 |
0.3 |
GO:0046855 |
phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 |
0.1 |
GO:0006817 |
phosphate ion transport(GO:0006817) |
| 0.0 |
0.1 |
GO:0007202 |
activation of phospholipase C activity(GO:0007202) |
| 0.0 |
0.0 |
GO:0060218 |
hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 |
0.1 |
GO:0051084 |
'de novo' protein folding(GO:0006458) 'de novo' posttranslational protein folding(GO:0051084) |
| 0.0 |
0.1 |
GO:0061470 |
interleukin-21 production(GO:0032625) T follicular helper cell differentiation(GO:0061470) interleukin-21 secretion(GO:0072619) |
| 0.0 |
0.1 |
GO:0019276 |
UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 |
0.0 |
GO:0050819 |
negative regulation of coagulation(GO:0050819) |
| 0.0 |
0.1 |
GO:0089711 |
L-glutamate transmembrane transport(GO:0089711) |
| 0.0 |
1.1 |
GO:0051028 |
mRNA transport(GO:0051028) |
| 0.0 |
0.5 |
GO:0070527 |
platelet aggregation(GO:0070527) |
| 0.0 |
0.1 |
GO:0006123 |
mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 |
0.3 |
GO:0043392 |
negative regulation of DNA binding(GO:0043392) |
| 0.0 |
0.0 |
GO:0048208 |
vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.0 |
0.1 |
GO:0072337 |
modified amino acid transport(GO:0072337) |
| 0.0 |
0.2 |
GO:0030201 |
heparan sulfate proteoglycan metabolic process(GO:0030201) |
| 0.0 |
0.2 |
GO:0045662 |
negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 |
0.3 |
GO:0033120 |
positive regulation of RNA splicing(GO:0033120) |
| 0.0 |
0.1 |
GO:1904587 |
glycoprotein ERAD pathway(GO:0097466) response to glycoprotein(GO:1904587) |
| 0.0 |
0.2 |
GO:0043171 |
peptide catabolic process(GO:0043171) |
| 0.0 |
0.2 |
GO:0050982 |
detection of mechanical stimulus(GO:0050982) |
| 0.0 |
0.4 |
GO:2000648 |
positive regulation of stem cell proliferation(GO:2000648) |
| 0.0 |
0.5 |
GO:0000077 |
DNA damage checkpoint(GO:0000077) DNA integrity checkpoint(GO:0031570) |
| 0.0 |
0.1 |
GO:0019886 |
antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.0 |
0.1 |
GO:0048745 |
smooth muscle tissue development(GO:0048745) |
| 0.0 |
0.1 |
GO:0050818 |
regulation of coagulation(GO:0050818) |
| 0.0 |
0.3 |
GO:0060968 |
regulation of gene silencing(GO:0060968) |
| 0.0 |
0.1 |
GO:1902230 |
negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.0 |
2.2 |
GO:0008380 |
RNA splicing(GO:0008380) |