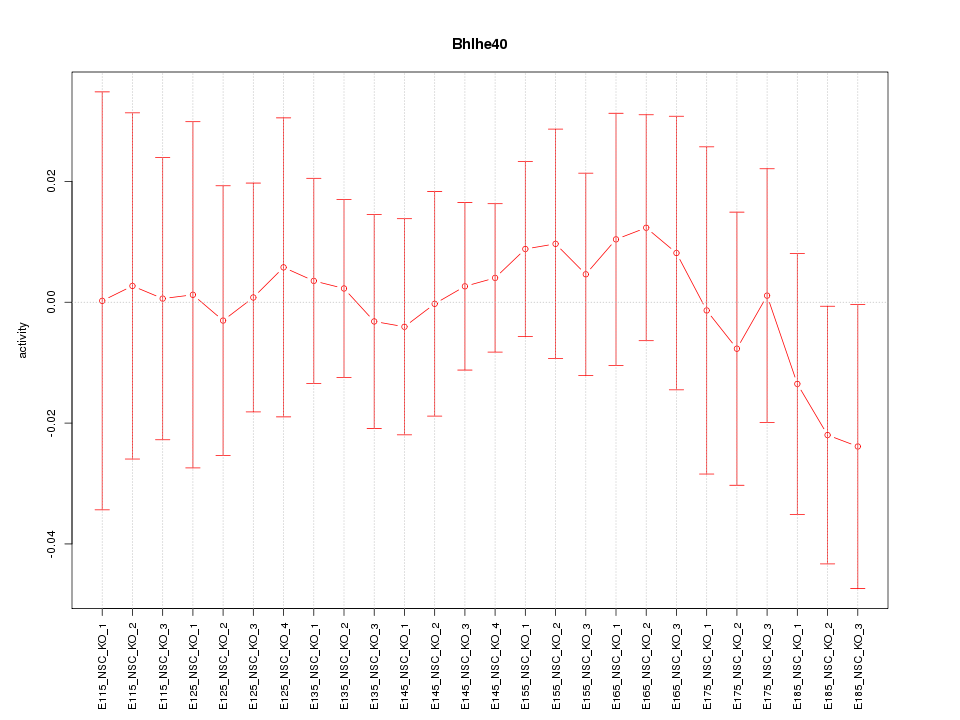
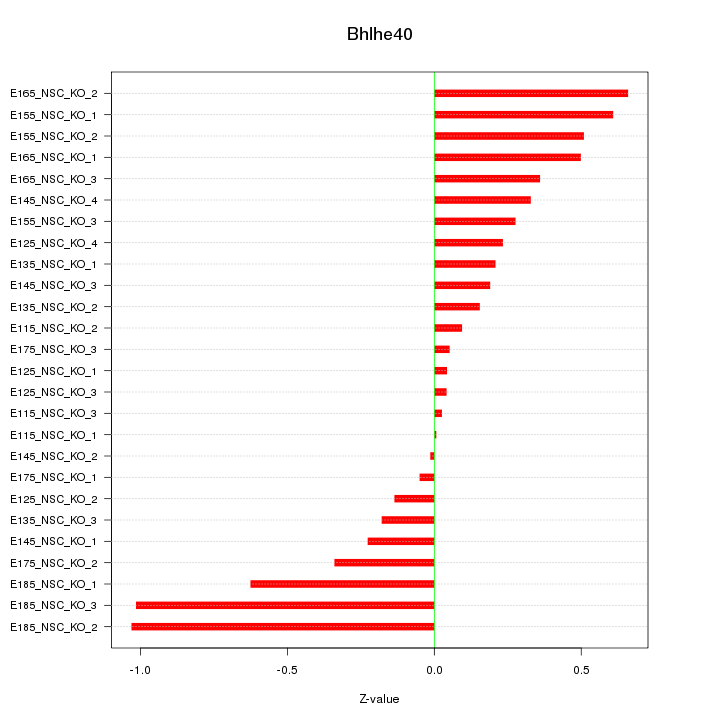
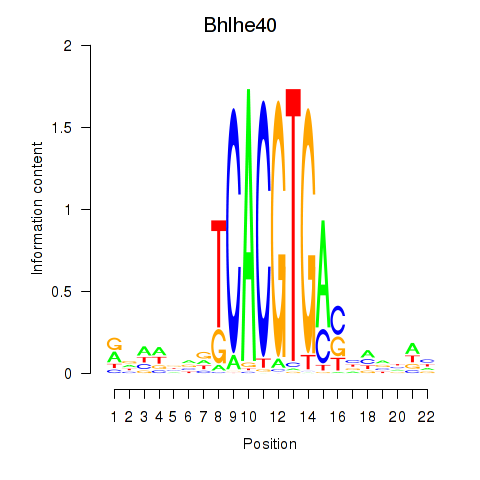
Motif ID: Bhlhe40
Z-value: 0.416
Transcription factors associated with Bhlhe40:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Bhlhe40 | ENSMUSG00000030103.5 | Bhlhe40 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Bhlhe40 | mm10_v2_chr6_+_108660772_108660874 | -0.50 | 9.0e-03 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.5 | GO:0070829 | heterochromatin maintenance(GO:0070829) |
| 0.1 | 0.3 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.1 | 0.3 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.1 | 0.2 | GO:0002940 | tRNA N2-guanine methylation(GO:0002940) |
| 0.1 | 0.2 | GO:1902219 | regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902218) negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.1 | 0.3 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.1 | 0.2 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.1 | 0.2 | GO:0006407 | rRNA export from nucleus(GO:0006407) |
| 0.1 | 0.2 | GO:0046032 | ADP catabolic process(GO:0046032) |
| 0.0 | 0.1 | GO:2000508 | regulation of dendritic cell chemotaxis(GO:2000508) positive regulation of dendritic cell chemotaxis(GO:2000510) |
| 0.0 | 0.4 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.4 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 | 0.2 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.0 | 0.2 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 0.2 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.0 | 0.1 | GO:0002922 | positive regulation of humoral immune response(GO:0002922) progesterone receptor signaling pathway(GO:0050847) |
| 0.0 | 0.2 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.7 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.0 | 0.6 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.3 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.0 | 0.3 | GO:0080111 | DNA demethylation(GO:0080111) |
| 0.0 | 0.1 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.3 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.1 | GO:0035542 | endosomal vesicle fusion(GO:0034058) regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.2 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.4 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.2 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.1 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 0.1 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.1 | 0.4 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 0.3 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.7 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.1 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.2 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.2 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.6 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.1 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.3 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 0.1 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.1 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.1 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.1 | 0.3 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.1 | 0.3 | GO:0008311 | double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 0.1 | 0.4 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.1 | 0.7 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.1 | 0.2 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.1 | 0.2 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.1 | 0.2 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.1 | 0.2 | GO:0015173 | hydrogen:amino acid symporter activity(GO:0005280) aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.1 | 0.2 | GO:0000832 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.0 | 0.3 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.1 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.0 | 0.1 | GO:0001851 | complement component C3b binding(GO:0001851) proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.8 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.6 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.2 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.2 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.2 | GO:0047631 | adenosine-diphosphatase activity(GO:0043262) ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.2 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.4 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.1 | GO:0016936 | galactoside binding(GO:0016936) |