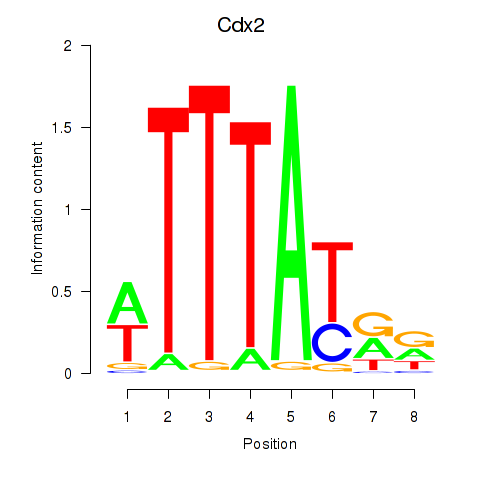
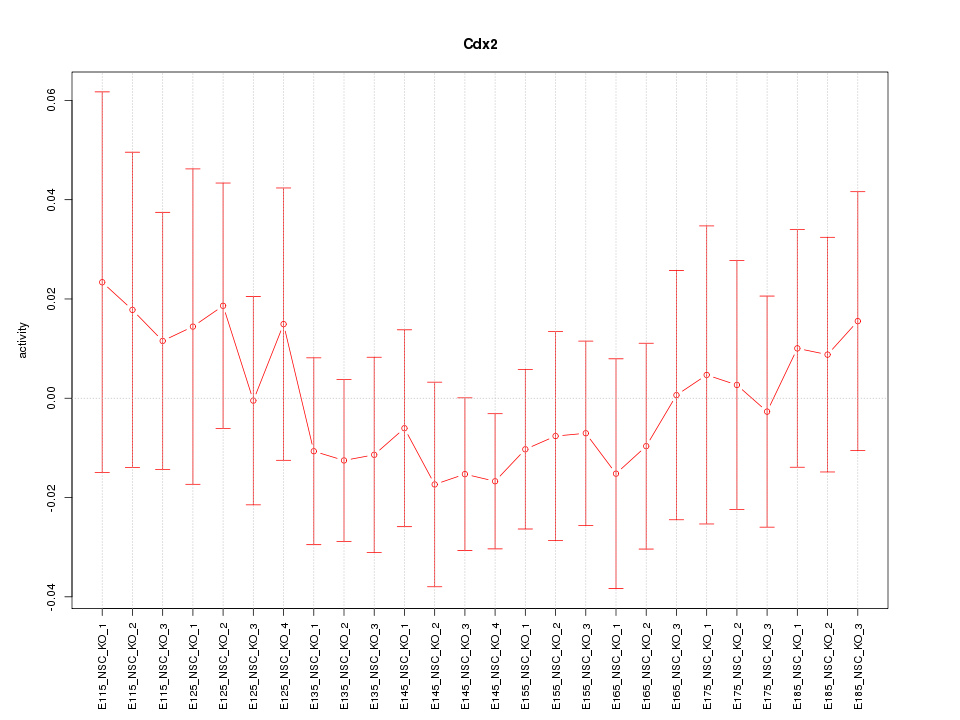
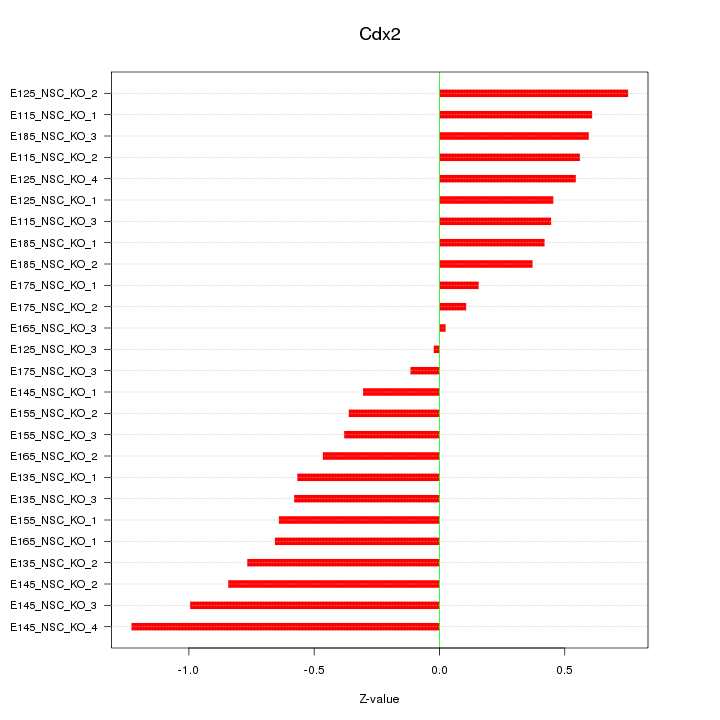
| 0.5 |
3.2 |
GO:0021555 |
midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.3 |
1.0 |
GO:0014846 |
esophagus smooth muscle contraction(GO:0014846) |
| 0.2 |
0.9 |
GO:0035606 |
peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.2 |
1.5 |
GO:0001887 |
selenium compound metabolic process(GO:0001887) |
| 0.2 |
0.9 |
GO:0035022 |
positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.2 |
0.5 |
GO:0046368 |
GDP-L-fucose metabolic process(GO:0046368) |
| 0.2 |
0.5 |
GO:0042320 |
regulation of circadian sleep/wake cycle, REM sleep(GO:0042320) circadian sleep/wake cycle, REM sleep(GO:0042747) positive regulation of circadian sleep/wake cycle, sleep(GO:0045938) |
| 0.1 |
0.8 |
GO:0021913 |
regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.1 |
0.5 |
GO:1903887 |
motile primary cilium assembly(GO:1903887) |
| 0.1 |
1.7 |
GO:0006825 |
copper ion transport(GO:0006825) |
| 0.1 |
0.4 |
GO:0030644 |
cellular chloride ion homeostasis(GO:0030644) |
| 0.1 |
0.3 |
GO:0001550 |
ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.1 |
0.5 |
GO:0019227 |
neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 |
0.6 |
GO:0048664 |
neuron fate determination(GO:0048664) |
| 0.0 |
0.6 |
GO:0015937 |
coenzyme A biosynthetic process(GO:0015937) |
| 0.0 |
0.1 |
GO:0048818 |
regulation of integrin biosynthetic process(GO:0045113) positive regulation of hair follicle maturation(GO:0048818) positive regulation of catagen(GO:0051795) regulation of epithelial to mesenchymal transition involved in endocardial cushion formation(GO:1905005) |
| 0.0 |
0.3 |
GO:0060665 |
regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 |
0.2 |
GO:0035590 |
purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.0 |
0.6 |
GO:0032291 |
central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.0 |
0.2 |
GO:0038018 |
Wnt receptor catabolic process(GO:0038018) |
| 0.0 |
0.1 |
GO:0090135 |
actin filament branching(GO:0090135) negative regulation of phospholipase C activity(GO:1900275) |
| 0.0 |
0.5 |
GO:1902043 |
positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.0 |
0.4 |
GO:0060391 |
positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 |
0.2 |
GO:0060693 |
regulation of branching involved in salivary gland morphogenesis(GO:0060693) |
| 0.0 |
0.2 |
GO:0035994 |
response to muscle stretch(GO:0035994) |
| 0.0 |
0.1 |
GO:0030263 |
apoptotic chromosome condensation(GO:0030263) |
| 0.0 |
0.1 |
GO:0071285 |
cellular response to lithium ion(GO:0071285) |