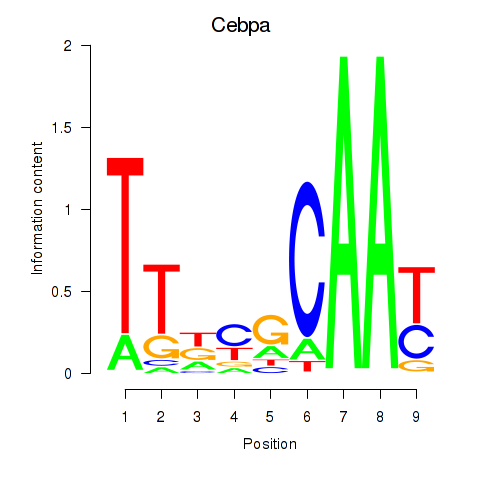
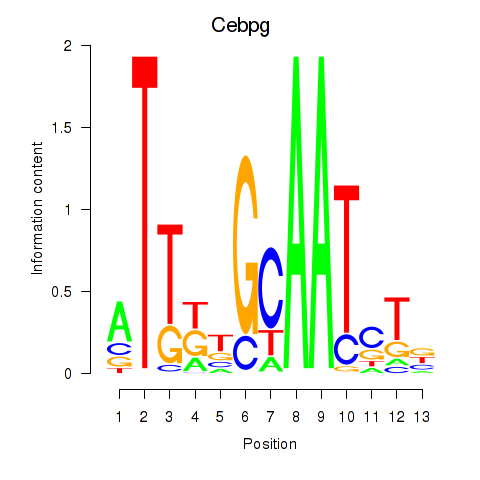
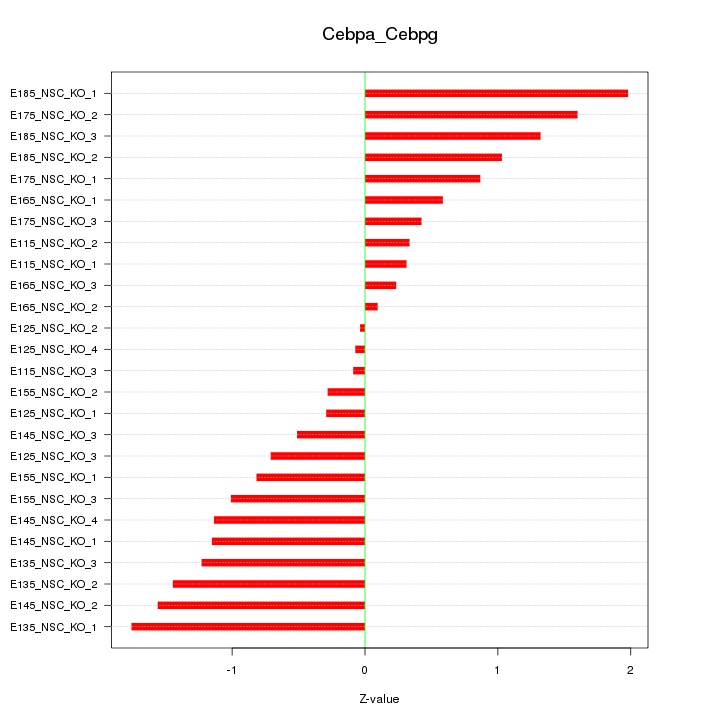
| 1.9 |
11.6 |
GO:1902847 |
macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of tau-protein kinase activity(GO:1902949) |
| 1.0 |
3.1 |
GO:0045410 |
positive regulation of interleukin-6 biosynthetic process(GO:0045410) |
| 1.0 |
2.9 |
GO:0008228 |
opsonization(GO:0008228) modulation of molecular function in other organism(GO:0044359) negative regulation of molecular function in other organism(GO:0044362) negative regulation of molecular function in other organism involved in symbiotic interaction(GO:0052204) modulation of molecular function in other organism involved in symbiotic interaction(GO:0052205) negative regulation by host of symbiont molecular function(GO:0052405) modification by host of symbiont molecular function(GO:0052428) |
| 0.5 |
1.4 |
GO:0032430 |
positive regulation of phospholipase A2 activity(GO:0032430) |
| 0.5 |
4.1 |
GO:0060665 |
regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.4 |
1.7 |
GO:0010808 |
positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.4 |
2.0 |
GO:0046684 |
response to pyrethroid(GO:0046684) |
| 0.3 |
1.0 |
GO:0002586 |
positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002586) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) detection of peptidoglycan(GO:0032499) |
| 0.3 |
1.2 |
GO:2000427 |
positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.3 |
1.8 |
GO:0070779 |
D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.2 |
2.4 |
GO:0051901 |
positive regulation of mitochondrial depolarization(GO:0051901) |
| 0.2 |
1.2 |
GO:0047484 |
regulation of response to osmotic stress(GO:0047484) |
| 0.2 |
1.1 |
GO:0042167 |
heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.2 |
1.4 |
GO:0032264 |
IMP salvage(GO:0032264) |
| 0.2 |
3.0 |
GO:0046688 |
response to copper ion(GO:0046688) |
| 0.1 |
0.4 |
GO:0070944 |
neutrophil mediated killing of bacterium(GO:0070944) |
| 0.1 |
0.4 |
GO:0003278 |
apoptotic process involved in heart morphogenesis(GO:0003278) |
| 0.1 |
1.0 |
GO:1990118 |
sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 |
0.8 |
GO:0030643 |
cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.1 |
0.6 |
GO:0042701 |
progesterone secretion(GO:0042701) |
| 0.1 |
0.3 |
GO:1903903 |
regulation of establishment of T cell polarity(GO:1903903) |
| 0.1 |
0.6 |
GO:2000049 |
positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.1 |
0.3 |
GO:0090258 |
negative regulation of mitochondrial fission(GO:0090258) |
| 0.1 |
0.3 |
GO:0014877 |
response to inactivity(GO:0014854) response to muscle inactivity(GO:0014870) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.1 |
0.3 |
GO:2001206 |
positive regulation of osteoclast development(GO:2001206) |
| 0.1 |
0.9 |
GO:0010650 |
positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.1 |
0.5 |
GO:1902261 |
positive regulation of delayed rectifier potassium channel activity(GO:1902261) positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.1 |
0.4 |
GO:0051684 |
maintenance of Golgi location(GO:0051684) |
| 0.1 |
0.6 |
GO:0005513 |
detection of calcium ion(GO:0005513) |
| 0.1 |
0.3 |
GO:0019287 |
isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.1 |
0.1 |
GO:0002309 |
T cell proliferation involved in immune response(GO:0002309) |
| 0.1 |
0.3 |
GO:0097527 |
necroptotic signaling pathway(GO:0097527) |
| 0.1 |
0.5 |
GO:0050908 |
detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 |
0.4 |
GO:0071420 |
cellular response to histamine(GO:0071420) |
| 0.0 |
1.3 |
GO:0071398 |
cellular response to fatty acid(GO:0071398) |
| 0.0 |
1.4 |
GO:0032728 |
positive regulation of interferon-beta production(GO:0032728) |
| 0.0 |
0.4 |
GO:0006983 |
ER overload response(GO:0006983) |
| 0.0 |
0.3 |
GO:0070458 |
detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 0.0 |
0.1 |
GO:0002184 |
cytoplasmic translational termination(GO:0002184) |
| 0.0 |
1.6 |
GO:0045744 |
negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.0 |
1.9 |
GO:0008542 |
visual learning(GO:0008542) |
| 0.0 |
0.2 |
GO:1903441 |
protein localization to ciliary membrane(GO:1903441) |
| 0.0 |
0.1 |
GO:0070782 |
phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.0 |
0.5 |
GO:0050829 |
defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 |
0.3 |
GO:1901898 |
negative regulation of relaxation of muscle(GO:1901078) regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 |
0.1 |
GO:0016557 |
peroxisome membrane biogenesis(GO:0016557) |
| 0.0 |
0.1 |
GO:1901165 |
positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 |
0.1 |
GO:0018364 |
peptidyl-glutamine methylation(GO:0018364) |
| 0.0 |
0.4 |
GO:0030033 |
microvillus assembly(GO:0030033) |
| 0.0 |
0.1 |
GO:0046898 |
response to cycloheximide(GO:0046898) |
| 0.0 |
0.2 |
GO:0016127 |
cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 |
0.1 |
GO:1902309 |
negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.0 |
0.3 |
GO:0010758 |
regulation of macrophage chemotaxis(GO:0010758) |
| 0.0 |
0.3 |
GO:0006828 |
manganese ion transport(GO:0006828) |
| 0.0 |
0.6 |
GO:1900449 |
regulation of glutamate receptor signaling pathway(GO:1900449) |
| 0.0 |
0.3 |
GO:0007340 |
acrosome reaction(GO:0007340) |
| 0.0 |
0.4 |
GO:0007608 |
sensory perception of smell(GO:0007608) |
| 0.0 |
0.4 |
GO:0051497 |
negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 |
0.2 |
GO:0030150 |
protein import into mitochondrial matrix(GO:0030150) |
| 0.0 |
1.6 |
GO:0007050 |
cell cycle arrest(GO:0007050) |
| 0.0 |
0.0 |
GO:0006421 |
asparaginyl-tRNA aminoacylation(GO:0006421) |