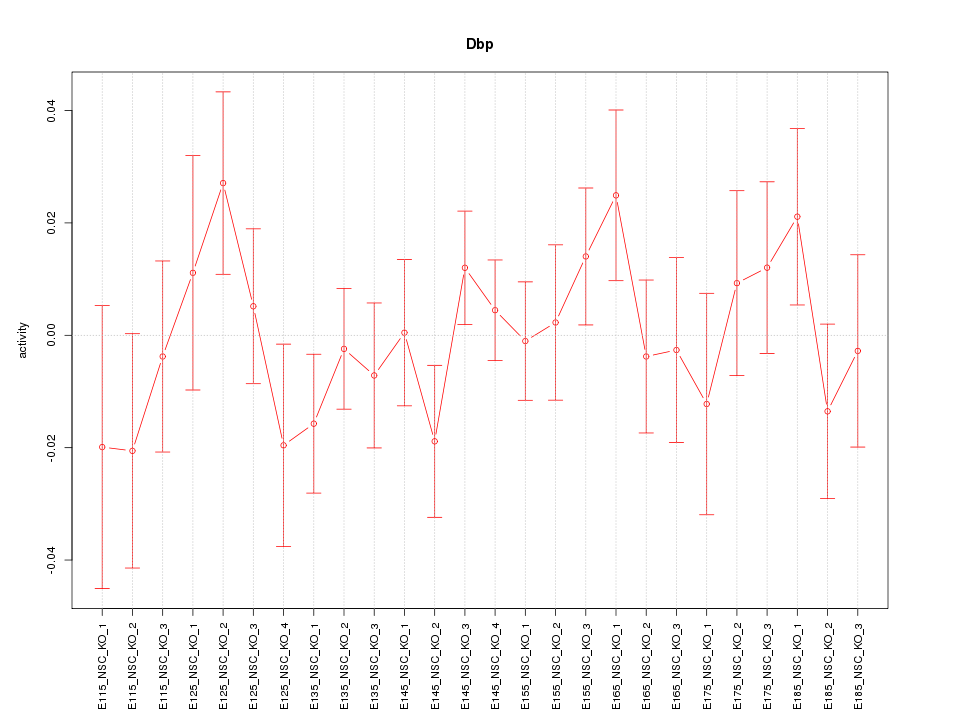
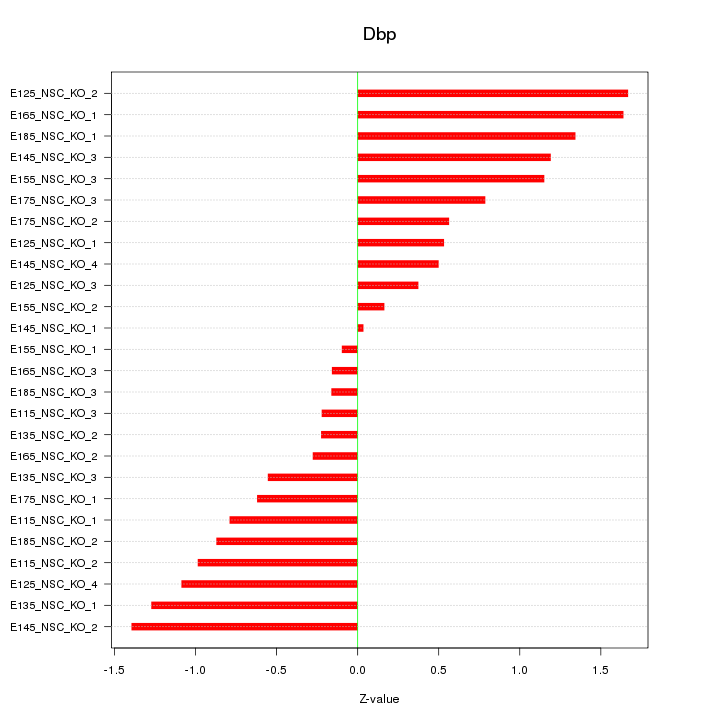
| 1.0 |
3.0 |
GO:0001928 |
regulation of exocyst assembly(GO:0001928) |
| 0.4 |
2.1 |
GO:2000302 |
regulation of synaptic vesicle endocytosis(GO:1900242) positive regulation of synaptic vesicle exocytosis(GO:2000302) |
| 0.4 |
1.1 |
GO:0086046 |
membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.3 |
1.7 |
GO:0045759 |
negative regulation of action potential(GO:0045759) |
| 0.3 |
1.1 |
GO:1901204 |
positive regulation of guanylate cyclase activity(GO:0031284) regulation of adrenergic receptor signaling pathway involved in heart process(GO:1901204) |
| 0.3 |
2.3 |
GO:0014824 |
artery smooth muscle contraction(GO:0014824) |
| 0.2 |
2.9 |
GO:1900383 |
regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.2 |
2.1 |
GO:0071420 |
cellular response to histamine(GO:0071420) |
| 0.2 |
2.0 |
GO:0035887 |
aortic smooth muscle cell differentiation(GO:0035887) |
| 0.2 |
2.0 |
GO:0090043 |
regulation of tubulin deacetylation(GO:0090043) |
| 0.2 |
1.7 |
GO:0061368 |
behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.2 |
1.3 |
GO:0060040 |
retinal bipolar neuron differentiation(GO:0060040) |
| 0.2 |
0.6 |
GO:0019482 |
beta-alanine metabolic process(GO:0019482) |
| 0.2 |
0.9 |
GO:0071205 |
regulation of axon diameter(GO:0031133) clustering of voltage-gated potassium channels(GO:0045163) protein localization to juxtaparanode region of axon(GO:0071205) |
| 0.2 |
0.6 |
GO:0021893 |
cerebral cortex GABAergic interneuron fate commitment(GO:0021893) |
| 0.2 |
1.1 |
GO:0007258 |
JUN phosphorylation(GO:0007258) |
| 0.2 |
1.1 |
GO:0098700 |
aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.2 |
1.1 |
GO:0089711 |
L-glutamate transmembrane transport(GO:0089711) |
| 0.2 |
1.2 |
GO:0071361 |
cellular response to zinc ion(GO:0071294) cellular response to ethanol(GO:0071361) |
| 0.2 |
0.5 |
GO:0032241 |
positive regulation of nucleobase-containing compound transport(GO:0032241) regulation of nucleoside transport(GO:0032242) negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) negative regulation of mucus secretion(GO:0070256) |
| 0.2 |
1.0 |
GO:2000672 |
negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.2 |
1.3 |
GO:0097264 |
self proteolysis(GO:0097264) |
| 0.2 |
1.2 |
GO:0048625 |
myoblast fate commitment(GO:0048625) |
| 0.2 |
0.5 |
GO:0003289 |
atrial septum primum morphogenesis(GO:0003289) ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 0.2 |
0.6 |
GO:2000211 |
regulation of glutamate metabolic process(GO:2000211) |
| 0.2 |
2.6 |
GO:2001014 |
regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.1 |
0.3 |
GO:0016115 |
terpenoid catabolic process(GO:0016115) |
| 0.1 |
0.4 |
GO:2000569 |
T-helper 2 cell activation(GO:0035712) positive regulation of T-helper 17 type immune response(GO:2000318) regulation of T-helper 2 cell activation(GO:2000569) positive regulation of T-helper 2 cell activation(GO:2000570) |
| 0.1 |
1.1 |
GO:0050908 |
detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.1 |
0.4 |
GO:0050883 |
musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.1 |
1.6 |
GO:0021540 |
corpus callosum morphogenesis(GO:0021540) |
| 0.1 |
1.6 |
GO:0009312 |
oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 |
0.8 |
GO:0009449 |
gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.1 |
0.6 |
GO:0060050 |
positive regulation of protein glycosylation(GO:0060050) |
| 0.1 |
2.2 |
GO:0035235 |
ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.1 |
0.4 |
GO:1990743 |
protein sialylation(GO:1990743) |
| 0.1 |
1.0 |
GO:1902018 |
negative regulation of cilium assembly(GO:1902018) |
| 0.1 |
0.9 |
GO:0007379 |
segment specification(GO:0007379) |
| 0.1 |
0.2 |
GO:1900186 |
negative regulation of clathrin-mediated endocytosis(GO:1900186) |
| 0.1 |
0.4 |
GO:0071415 |
cellular response to caffeine(GO:0071313) cellular response to purine-containing compound(GO:0071415) |
| 0.1 |
1.8 |
GO:0043552 |
positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.1 |
1.1 |
GO:0010758 |
regulation of macrophage chemotaxis(GO:0010758) |
| 0.1 |
0.9 |
GO:1901898 |
negative regulation of relaxation of muscle(GO:1901078) regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 |
0.6 |
GO:0016338 |
calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 |
0.2 |
GO:0048686 |
regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 0.1 |
1.2 |
GO:0008090 |
retrograde axonal transport(GO:0008090) |
| 0.1 |
1.0 |
GO:1902993 |
positive regulation of beta-amyloid formation(GO:1902004) positive regulation of amyloid precursor protein catabolic process(GO:1902993) |
| 0.1 |
2.3 |
GO:0051491 |
positive regulation of filopodium assembly(GO:0051491) |
| 0.1 |
0.3 |
GO:0050861 |
positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.1 |
0.7 |
GO:0021559 |
trigeminal nerve development(GO:0021559) |
| 0.1 |
0.4 |
GO:0015015 |
heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 |
0.3 |
GO:0051964 |
negative regulation of synapse assembly(GO:0051964) |
| 0.1 |
1.4 |
GO:0032011 |
ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 |
0.6 |
GO:0051103 |
DNA ligation involved in DNA repair(GO:0051103) |
| 0.1 |
0.3 |
GO:0006287 |
base-excision repair, gap-filling(GO:0006287) |
| 0.1 |
0.3 |
GO:0045607 |
regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.1 |
0.6 |
GO:0048266 |
behavioral response to pain(GO:0048266) |
| 0.1 |
4.1 |
GO:0010923 |
negative regulation of phosphatase activity(GO:0010923) |
| 0.1 |
0.3 |
GO:0021914 |
negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.1 |
1.3 |
GO:0070306 |
lens fiber cell differentiation(GO:0070306) |
| 0.0 |
0.1 |
GO:0045014 |
detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) detection of glucose(GO:0051594) |
| 0.0 |
0.9 |
GO:0010669 |
epithelial structure maintenance(GO:0010669) |
| 0.0 |
0.1 |
GO:0021529 |
spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.0 |
1.0 |
GO:0051968 |
positive regulation of synaptic transmission, glutamatergic(GO:0051968) |
| 0.0 |
0.3 |
GO:0030828 |
positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.0 |
0.4 |
GO:0048672 |
positive regulation of collateral sprouting(GO:0048672) |
| 0.0 |
0.8 |
GO:0048026 |
positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 |
1.4 |
GO:0032728 |
positive regulation of interferon-beta production(GO:0032728) |
| 0.0 |
0.5 |
GO:0021527 |
spinal cord association neuron differentiation(GO:0021527) |
| 0.0 |
0.2 |
GO:0036324 |
negative regulation of toll-like receptor 4 signaling pathway(GO:0034144) vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 0.0 |
0.2 |
GO:1902969 |
mitotic DNA replication(GO:1902969) |
| 0.0 |
0.1 |
GO:0097105 |
presynaptic membrane assembly(GO:0097105) |
| 0.0 |
0.6 |
GO:0046069 |
cGMP catabolic process(GO:0046069) |
| 0.0 |
0.4 |
GO:0060837 |
blood vessel endothelial cell differentiation(GO:0060837) |
| 0.0 |
0.3 |
GO:1901621 |
regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901620) negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 |
0.3 |
GO:1900042 |
positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.0 |
0.4 |
GO:0060219 |
camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.0 |
0.2 |
GO:0071476 |
cellular hypotonic response(GO:0071476) |
| 0.0 |
0.2 |
GO:0010993 |
regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 0.0 |
0.2 |
GO:0006344 |
maintenance of chromatin silencing(GO:0006344) |
| 0.0 |
0.5 |
GO:0007097 |
nuclear migration(GO:0007097) |
| 0.0 |
0.9 |
GO:0051452 |
intracellular pH reduction(GO:0051452) |
| 0.0 |
0.3 |
GO:0007250 |
activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 |
0.6 |
GO:0090036 |
regulation of protein kinase C signaling(GO:0090036) |
| 0.0 |
0.2 |
GO:0046548 |
retinal rod cell development(GO:0046548) |
| 0.0 |
0.2 |
GO:0030718 |
germ-line stem cell population maintenance(GO:0030718) |
| 0.0 |
1.1 |
GO:0034605 |
cellular response to heat(GO:0034605) |
| 0.0 |
0.1 |
GO:0035609 |
C-terminal protein deglutamylation(GO:0035609) |
| 0.0 |
1.1 |
GO:0030901 |
midbrain development(GO:0030901) |
| 0.0 |
1.8 |
GO:0007156 |
homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 |
0.8 |
GO:0045214 |
sarcomere organization(GO:0045214) |
| 0.0 |
0.1 |
GO:0036438 |
maintenance of lens transparency(GO:0036438) |
| 0.0 |
0.5 |
GO:0060043 |
regulation of cardiac muscle cell proliferation(GO:0060043) |
| 0.0 |
0.4 |
GO:0071625 |
vocalization behavior(GO:0071625) |
| 0.0 |
1.4 |
GO:0060395 |
SMAD protein signal transduction(GO:0060395) |
| 0.0 |
0.6 |
GO:0006491 |
N-glycan processing(GO:0006491) |
| 0.0 |
0.1 |
GO:0035063 |
nuclear speck organization(GO:0035063) |
| 0.0 |
1.7 |
GO:0001578 |
microtubule bundle formation(GO:0001578) |
| 0.0 |
2.3 |
GO:0098792 |
xenophagy(GO:0098792) |
| 0.0 |
0.2 |
GO:0021684 |
cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 |
0.2 |
GO:0060038 |
cardiac muscle cell proliferation(GO:0060038) |
| 0.0 |
1.2 |
GO:0050775 |
positive regulation of dendrite morphogenesis(GO:0050775) |
| 0.0 |
0.2 |
GO:0043982 |
histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 |
0.7 |
GO:0006376 |
mRNA splice site selection(GO:0006376) |
| 0.0 |
0.2 |
GO:0021702 |
cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 |
0.5 |
GO:0048741 |
skeletal muscle fiber development(GO:0048741) |
| 0.0 |
0.1 |
GO:0019065 |
receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.0 |
0.3 |
GO:0071425 |
hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 |
0.3 |
GO:0000301 |
retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 |
1.0 |
GO:0030968 |
endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 |
0.2 |
GO:0071447 |
cellular response to hydroperoxide(GO:0071447) |
| 0.0 |
0.2 |
GO:0007214 |
gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 |
0.0 |
GO:0097155 |
fasciculation of sensory neuron axon(GO:0097155) |
| 0.0 |
0.4 |
GO:0046326 |
positive regulation of glucose import(GO:0046326) |
| 0.0 |
0.5 |
GO:0007200 |
phospholipase C-activating G-protein coupled receptor signaling pathway(GO:0007200) |
| 0.0 |
0.5 |
GO:0007173 |
epidermal growth factor receptor signaling pathway(GO:0007173) |