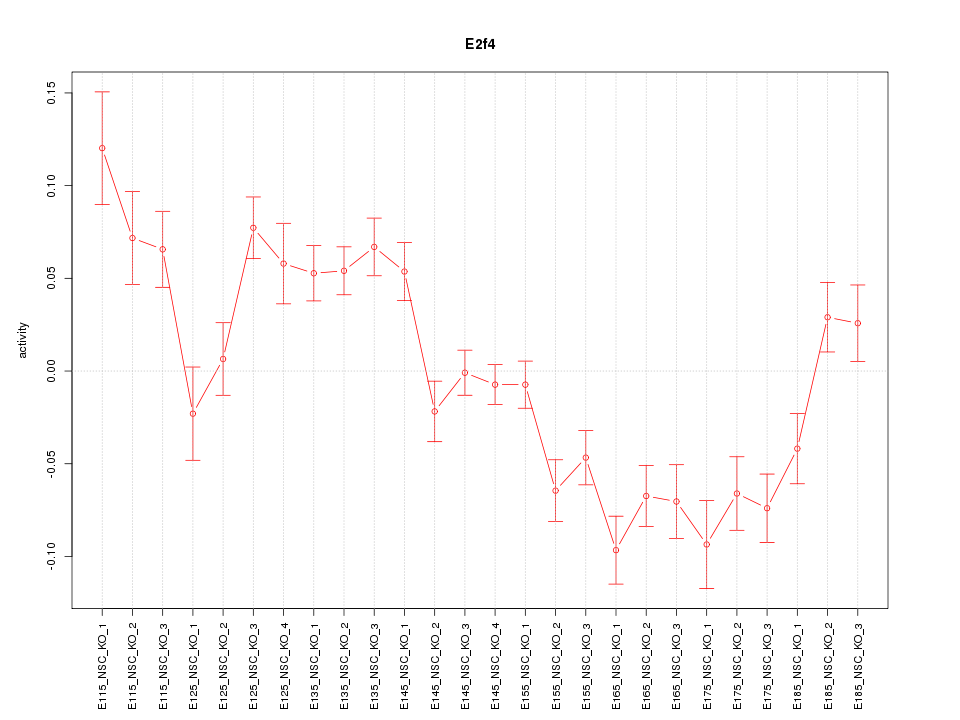
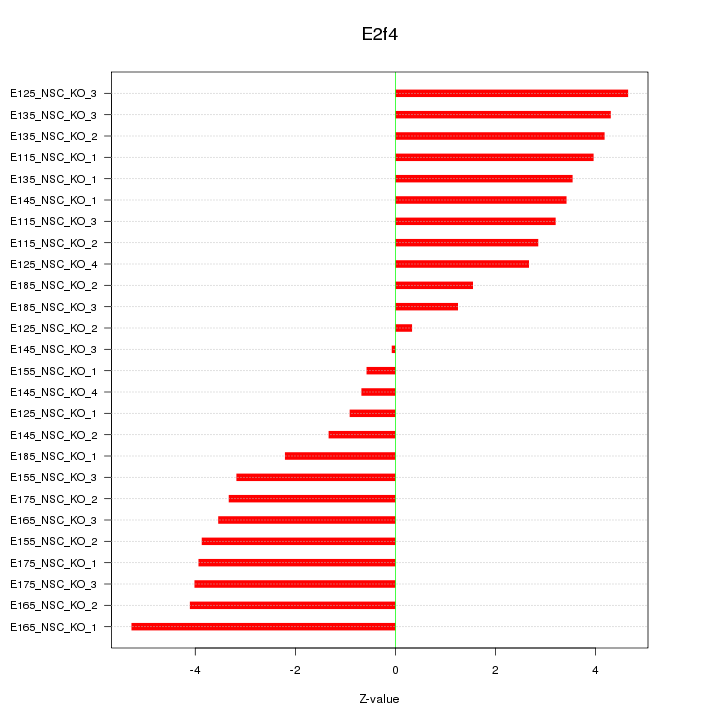
Motif ID: E2f4
Z-value: 3.165

Transcription factors associated with E2f4:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| E2f4 | ENSMUSG00000014859.8 | E2f4 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| E2f4 | mm10_v2_chr8_+_105297663_105297742 | 0.88 | 3.7e-09 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.1 | 24.6 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 5.1 | 15.3 | GO:0071921 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 3.9 | 11.6 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 3.6 | 10.9 | GO:0000087 | mitotic M phase(GO:0000087) |
| 3.5 | 10.5 | GO:0072076 | hyaluranon cable assembly(GO:0036118) nephrogenic mesenchyme development(GO:0072076) negative regulation of glomerular mesangial cell proliferation(GO:0072125) negative regulation of glomerulus development(GO:0090194) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 2.6 | 18.0 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 2.5 | 7.6 | GO:0010911 | regulation of isomerase activity(GO:0010911) positive regulation of isomerase activity(GO:0010912) phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 2.4 | 7.2 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 2.2 | 6.5 | GO:0003219 | atrioventricular node development(GO:0003162) cardiac right ventricle formation(GO:0003219) |
| 2.1 | 6.2 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 1.7 | 24.2 | GO:0070986 | left/right axis specification(GO:0070986) |
| 1.6 | 6.5 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 1.6 | 3.2 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 1.6 | 4.7 | GO:0045410 | positive regulation of interleukin-6 biosynthetic process(GO:0045410) |
| 1.5 | 8.8 | GO:0007144 | female meiosis I(GO:0007144) |
| 1.4 | 9.9 | GO:0000320 | re-entry into mitotic cell cycle(GO:0000320) |
| 1.3 | 8.8 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 1.3 | 6.3 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 1.2 | 10.5 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 1.1 | 4.4 | GO:1902990 | mitotic telomere maintenance via semi-conservative replication(GO:1902990) negative regulation of t-circle formation(GO:1904430) |
| 1.0 | 11.1 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 1.0 | 7.9 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.9 | 20.0 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.9 | 2.7 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.9 | 7.2 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.9 | 4.4 | GO:1900170 | negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.9 | 3.5 | GO:0035878 | nail development(GO:0035878) |
| 0.8 | 3.2 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.8 | 2.4 | GO:0003349 | epicardium-derived cardiac endothelial cell differentiation(GO:0003349) |
| 0.8 | 2.3 | GO:0060821 | inactivation of X chromosome by DNA methylation(GO:0060821) |
| 0.8 | 23.5 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.7 | 5.2 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.7 | 2.2 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.7 | 2.1 | GO:1902915 | negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.7 | 5.3 | GO:0010804 | negative regulation of tumor necrosis factor-mediated signaling pathway(GO:0010804) |
| 0.6 | 4.3 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) hydrogen sulfide biosynthetic process(GO:0070814) |
| 0.6 | 3.6 | GO:0070561 | vitamin D receptor signaling pathway(GO:0070561) |
| 0.5 | 17.9 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.5 | 1.6 | GO:0043987 | histone H3-S10 phosphorylation(GO:0043987) |
| 0.5 | 5.2 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.5 | 13.1 | GO:0040034 | regulation of development, heterochronic(GO:0040034) |
| 0.5 | 12.3 | GO:0051310 | metaphase plate congression(GO:0051310) |
| 0.5 | 2.0 | GO:0070589 | cell wall mannoprotein biosynthetic process(GO:0000032) mannoprotein metabolic process(GO:0006056) mannoprotein biosynthetic process(GO:0006057) cell wall glycoprotein biosynthetic process(GO:0031506) cell wall biogenesis(GO:0042546) cell wall macromolecule metabolic process(GO:0044036) cell wall macromolecule biosynthetic process(GO:0044038) chain elongation of O-linked mannose residue(GO:0044845) cellular component macromolecule biosynthetic process(GO:0070589) cell wall organization or biogenesis(GO:0071554) |
| 0.5 | 4.9 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.5 | 2.0 | GO:0046125 | pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.5 | 41.9 | GO:0007052 | mitotic spindle organization(GO:0007052) |
| 0.4 | 5.3 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.4 | 10.4 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.4 | 4.7 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.4 | 2.1 | GO:0046909 | intermembrane transport(GO:0046909) |
| 0.3 | 6.3 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.3 | 3.9 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.3 | 4.5 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.3 | 16.4 | GO:0007051 | spindle organization(GO:0007051) |
| 0.3 | 5.0 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.3 | 7.9 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.3 | 1.7 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.3 | 4.1 | GO:0034508 | centromere complex assembly(GO:0034508) |
| 0.3 | 3.6 | GO:0009143 | nucleoside triphosphate catabolic process(GO:0009143) |
| 0.3 | 0.8 | GO:0039530 | MDA-5 signaling pathway(GO:0039530) |
| 0.3 | 1.8 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.3 | 3.5 | GO:0042640 | anagen(GO:0042640) |
| 0.2 | 6.1 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.2 | 8.6 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.2 | 0.8 | GO:1904008 | response to monosodium glutamate(GO:1904008) cellular response to monosodium glutamate(GO:1904009) |
| 0.2 | 2.8 | GO:0042790 | transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:0042790) |
| 0.2 | 8.7 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.2 | 1.9 | GO:0036123 | histone H3-K9 dimethylation(GO:0036123) |
| 0.2 | 1.3 | GO:0033184 | positive regulation of histone ubiquitination(GO:0033184) |
| 0.2 | 2.3 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.2 | 2.8 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.2 | 0.5 | GO:0060161 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) positive regulation of dopamine receptor signaling pathway(GO:0060161) |
| 0.2 | 2.3 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.1 | 1.0 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.1 | 4.9 | GO:0008333 | endosome to lysosome transport(GO:0008333) |
| 0.1 | 0.8 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) cellular lipid biosynthetic process(GO:0097384) ether biosynthetic process(GO:1901503) |
| 0.1 | 1.6 | GO:0033210 | leptin-mediated signaling pathway(GO:0033210) |
| 0.1 | 1.1 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.1 | 3.5 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 0.3 | GO:0031086 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.1 | 4.5 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.1 | 2.3 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.1 | 2.8 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.1 | 0.3 | GO:1903181 | regulation of dopamine biosynthetic process(GO:1903179) positive regulation of dopamine biosynthetic process(GO:1903181) |
| 0.1 | 0.9 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.1 | 3.5 | GO:0046580 | negative regulation of Ras protein signal transduction(GO:0046580) |
| 0.1 | 1.3 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.1 | 6.6 | GO:0000956 | nuclear-transcribed mRNA catabolic process(GO:0000956) |
| 0.1 | 7.4 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.1 | 0.3 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.1 | 0.4 | GO:0071955 | recycling endosome to Golgi transport(GO:0071955) |
| 0.1 | 0.2 | GO:0097155 | fasciculation of sensory neuron axon(GO:0097155) |
| 0.0 | 2.7 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 1.6 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.0 | 1.0 | GO:0006221 | pyrimidine nucleotide biosynthetic process(GO:0006221) |
| 0.0 | 1.2 | GO:0043330 | response to exogenous dsRNA(GO:0043330) |
| 0.0 | 0.2 | GO:0018377 | N-terminal protein myristoylation(GO:0006499) protein myristoylation(GO:0018377) |
| 0.0 | 0.4 | GO:0048714 | positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.0 | 1.4 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 1.4 | GO:0010212 | response to ionizing radiation(GO:0010212) |
| 0.0 | 2.4 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 6.9 | GO:0007067 | mitotic nuclear division(GO:0007067) |
| 0.0 | 1.0 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.9 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.3 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 2.1 | GO:0055085 | transmembrane transport(GO:0055085) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.9 | 23.5 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 2.2 | 10.8 | GO:0031523 | Myb complex(GO:0031523) |
| 1.7 | 15.3 | GO:0008278 | cohesin complex(GO:0008278) |
| 1.7 | 10.0 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 1.7 | 18.2 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 1.3 | 6.5 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 1.3 | 6.5 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 1.2 | 11.6 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 1.1 | 12.3 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 1.0 | 7.2 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 1.0 | 7.9 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.8 | 13.4 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.7 | 4.9 | GO:0070695 | FHF complex(GO:0070695) |
| 0.7 | 6.2 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.7 | 8.8 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.6 | 6.2 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.5 | 8.1 | GO:0042555 | MCM complex(GO:0042555) |
| 0.5 | 2.0 | GO:0031502 | dolichyl-phosphate-mannose-protein mannosyltransferase complex(GO:0031502) |
| 0.5 | 29.0 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.4 | 3.5 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.4 | 27.7 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.4 | 1.9 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.3 | 1.9 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.3 | 1.6 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.3 | 4.7 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.3 | 1.4 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.3 | 2.7 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.3 | 23.2 | GO:0000922 | spindle pole(GO:0000922) |
| 0.3 | 2.3 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.3 | 3.5 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.2 | 1.4 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.2 | 2.2 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.2 | 1.4 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.2 | 9.4 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.2 | 19.3 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.2 | 14.6 | GO:0005814 | centriole(GO:0005814) |
| 0.2 | 10.4 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.2 | 3.5 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 7.4 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.1 | 3.2 | GO:0030894 | replisome(GO:0030894) |
| 0.1 | 4.9 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.1 | 14.9 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.1 | 3.5 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 1.8 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.1 | 8.6 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 2.8 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 7.0 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.1 | 6.9 | GO:0005813 | centrosome(GO:0005813) |
| 0.1 | 1.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 0.5 | GO:0034709 | methylosome(GO:0034709) |
| 0.1 | 1.7 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 7.2 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 1.0 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 3.2 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 13.4 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.8 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 4.6 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 22.6 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 0.2 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 10.4 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 0.2 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 1.2 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.0 | 0.8 | GO:0019897 | extrinsic component of plasma membrane(GO:0019897) |
| 0.0 | 9.2 | GO:0031988 | membrane-bounded vesicle(GO:0031988) |
| 0.0 | 4.7 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.0 | 4.0 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 4.2 | GO:0005925 | focal adhesion(GO:0005925) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.4 | 31.1 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 4.1 | 24.6 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) lysophosphatidic acid receptor activity(GO:0070915) |
| 2.4 | 7.2 | GO:0071207 | histone pre-mRNA stem-loop binding(GO:0071207) |
| 2.1 | 6.2 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) single guanine insertion binding(GO:0032142) |
| 1.7 | 8.6 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 1.7 | 8.6 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 1.5 | 10.8 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 1.2 | 11.8 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 1.2 | 8.2 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 1.1 | 4.4 | GO:0038049 | glucocorticoid receptor activity(GO:0004883) transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 1.1 | 3.2 | GO:0001565 | phorbol ester receptor activity(GO:0001565) non-kinase phorbol ester receptor activity(GO:0001566) |
| 1.0 | 10.5 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.9 | 7.6 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.9 | 6.5 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.8 | 7.2 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.7 | 7.9 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.7 | 3.5 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.6 | 9.7 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.6 | 1.2 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.5 | 1.6 | GO:0035175 | histone kinase activity (H3-S10 specific)(GO:0035175) |
| 0.5 | 3.2 | GO:0052630 | CTP:2,3-di-O-geranylgeranyl-sn-glycero-1-phosphate cytidyltransferase activity(GO:0043338) phospholactate guanylyltransferase activity(GO:0043814) ATP:coenzyme F420 adenylyltransferase activity(GO:0043910) UDP-N-acetylgalactosamine diphosphorylase activity(GO:0052630) |
| 0.5 | 2.0 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.4 | 4.7 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.4 | 1.6 | GO:0097003 | adipokinetic hormone receptor activity(GO:0097003) |
| 0.4 | 10.4 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.3 | 2.7 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.3 | 10.9 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.3 | 2.3 | GO:0051718 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates(GO:0051718) |
| 0.3 | 2.0 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.3 | 12.3 | GO:0045502 | dynein binding(GO:0045502) |
| 0.3 | 1.0 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.3 | 5.0 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.3 | 1.3 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.2 | 3.6 | GO:0032552 | deoxyribonucleotide binding(GO:0032552) |
| 0.2 | 9.5 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.2 | 2.8 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.2 | 1.9 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.2 | 6.5 | GO:0005112 | Notch binding(GO:0005112) |
| 0.2 | 4.2 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.2 | 9.4 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.2 | 2.2 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.2 | 1.7 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.2 | 3.9 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.2 | 0.5 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.1 | 9.0 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.1 | 2.1 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 2.7 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 5.0 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.1 | 5.9 | GO:0045309 | protein phosphorylated amino acid binding(GO:0045309) |
| 0.1 | 2.3 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 1.1 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.1 | 5.1 | GO:0044824 | integrase activity(GO:0008907) T/G mismatch-specific endonuclease activity(GO:0043765) retroviral integrase activity(GO:0044823) retroviral 3' processing activity(GO:0044824) |
| 0.1 | 0.8 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.1 | 16.7 | GO:0004721 | phosphoprotein phosphatase activity(GO:0004721) |
| 0.1 | 14.3 | GO:0042393 | histone binding(GO:0042393) |
| 0.1 | 1.0 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 8.5 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.1 | 1.8 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.1 | 0.8 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.9 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 21.1 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 0.3 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.0 | 0.2 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 4.4 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 2.2 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 1.9 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 35.9 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.0 | 5.3 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 2.1 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 1.1 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.2 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.8 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 1.1 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.0 | 0.7 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.7 | GO:0042281 | dolichyl pyrophosphate Man9GlcNAc2 alpha-1,3-glucosyltransferase activity(GO:0042281) |