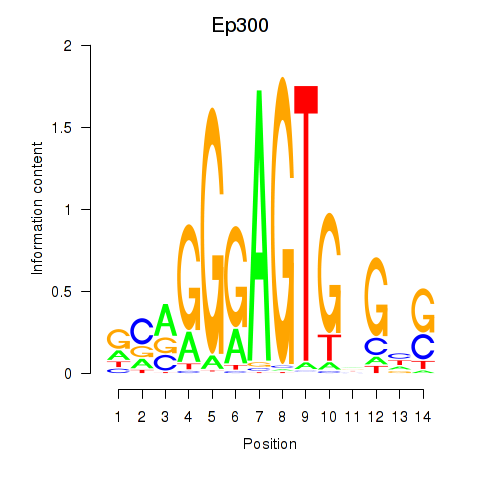
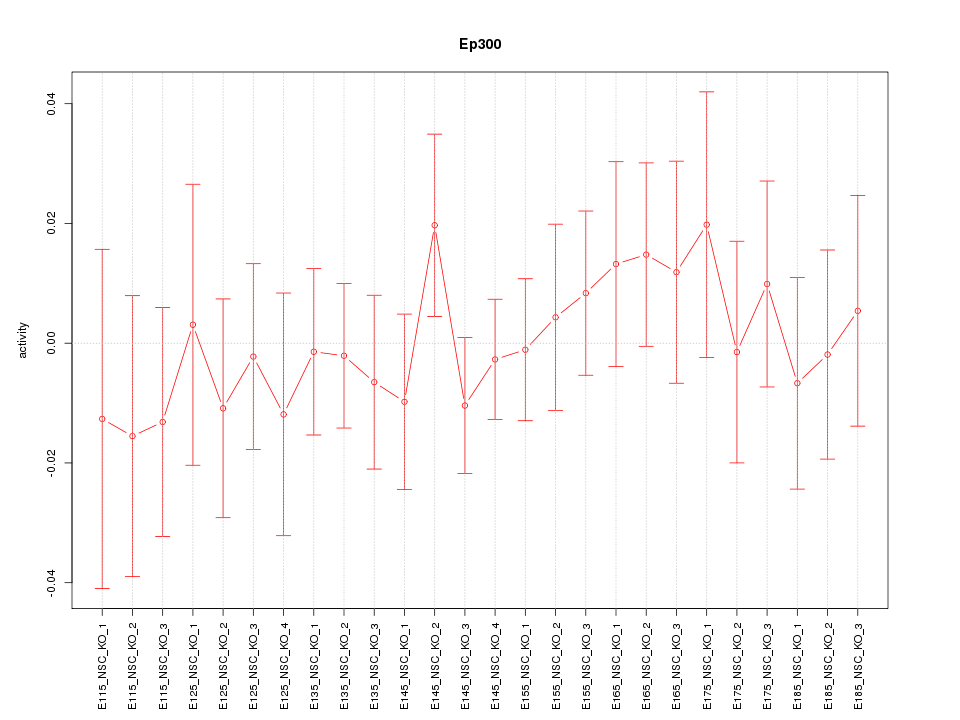
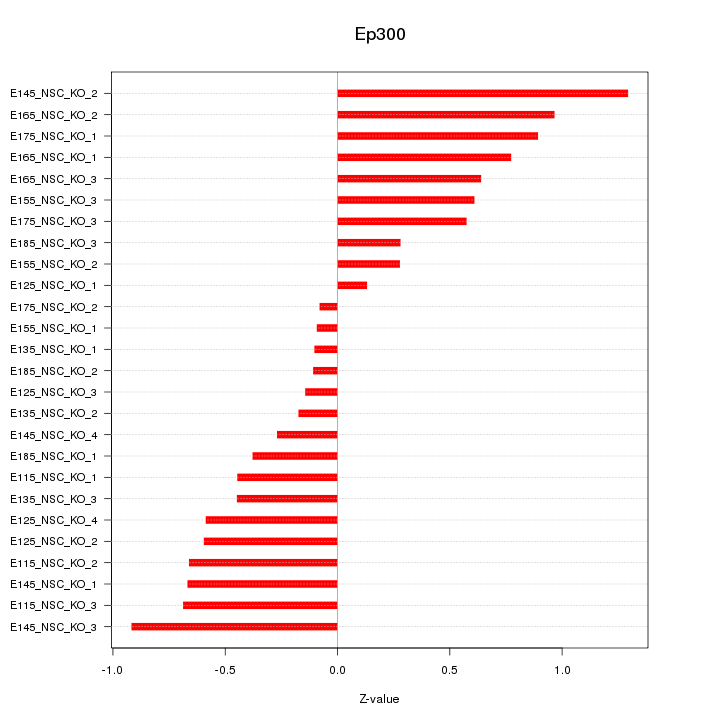
| 1.1 |
3.4 |
GO:0007621 |
negative regulation of female receptivity(GO:0007621) |
| 0.6 |
1.9 |
GO:1900736 |
regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900736) |
| 0.3 |
1.2 |
GO:0032289 |
central nervous system myelin formation(GO:0032289) cardiac cell fate specification(GO:0060912) |
| 0.2 |
2.0 |
GO:0005513 |
detection of calcium ion(GO:0005513) |
| 0.2 |
0.6 |
GO:0046122 |
purine deoxyribonucleoside metabolic process(GO:0046122) |
| 0.2 |
0.9 |
GO:0015803 |
branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.2 |
2.4 |
GO:0035635 |
entry of bacterium into host cell(GO:0035635) regulation of entry of bacterium into host cell(GO:2000535) |
| 0.2 |
0.7 |
GO:0010637 |
negative regulation of mitochondrial fusion(GO:0010637) |
| 0.2 |
2.2 |
GO:0071257 |
cellular response to electrical stimulus(GO:0071257) |
| 0.2 |
2.1 |
GO:0021684 |
cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.1 |
0.6 |
GO:0046898 |
response to cycloheximide(GO:0046898) |
| 0.1 |
0.5 |
GO:0034975 |
protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 |
0.5 |
GO:1903031 |
regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.1 |
2.3 |
GO:0014051 |
gamma-aminobutyric acid secretion(GO:0014051) |
| 0.1 |
0.7 |
GO:1904263 |
positive regulation of TORC1 signaling(GO:1904263) |
| 0.1 |
0.6 |
GO:0045347 |
negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.1 |
1.0 |
GO:0014049 |
positive regulation of glutamate secretion(GO:0014049) |
| 0.1 |
0.3 |
GO:1904154 |
trimming of terminal mannose on B branch(GO:0036509) positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.1 |
0.4 |
GO:0010936 |
negative regulation of macrophage cytokine production(GO:0010936) |
| 0.1 |
0.7 |
GO:1904217 |
regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) regulation of serine C-palmitoyltransferase activity(GO:1904220) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.1 |
0.4 |
GO:0060709 |
glycogen cell differentiation involved in embryonic placenta development(GO:0060709) |
| 0.1 |
0.7 |
GO:2000786 |
positive regulation of autophagosome assembly(GO:2000786) |
| 0.1 |
0.5 |
GO:0046880 |
regulation of follicle-stimulating hormone secretion(GO:0046880) follicle-stimulating hormone secretion(GO:0046884) |
| 0.1 |
0.3 |
GO:0071105 |
response to interleukin-11(GO:0071105) osteoclast fusion(GO:0072675) |
| 0.1 |
0.2 |
GO:0070172 |
positive regulation of tooth mineralization(GO:0070172) |
| 0.1 |
0.3 |
GO:0090148 |
membrane fission(GO:0090148) |
| 0.1 |
0.3 |
GO:2000298 |
regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.1 |
0.3 |
GO:0006049 |
UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.1 |
0.9 |
GO:0060837 |
blood vessel endothelial cell differentiation(GO:0060837) |
| 0.1 |
0.4 |
GO:1901837 |
negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.1 |
0.4 |
GO:0001955 |
blood vessel maturation(GO:0001955) |
| 0.1 |
0.4 |
GO:0090336 |
positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.1 |
0.5 |
GO:1902969 |
mitotic DNA replication(GO:1902969) |
| 0.0 |
0.1 |
GO:1901668 |
regulation of superoxide dismutase activity(GO:1901668) |
| 0.0 |
0.9 |
GO:0045793 |
positive regulation of cell size(GO:0045793) |
| 0.0 |
0.1 |
GO:0006669 |
sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.0 |
0.1 |
GO:0042732 |
D-xylose metabolic process(GO:0042732) |
| 0.0 |
0.5 |
GO:1901621 |
negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 |
0.7 |
GO:2000821 |
regulation of grooming behavior(GO:2000821) |
| 0.0 |
0.1 |
GO:0015960 |
diadenosine polyphosphate biosynthetic process(GO:0015960) diadenosine tetraphosphate metabolic process(GO:0015965) diadenosine tetraphosphate biosynthetic process(GO:0015966) |
| 0.0 |
0.3 |
GO:0051001 |
negative regulation of nitric-oxide synthase activity(GO:0051001) |
| 0.0 |
0.6 |
GO:0070935 |
3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 |
0.3 |
GO:0006183 |
GTP biosynthetic process(GO:0006183) |
| 0.0 |
0.2 |
GO:0006177 |
GMP biosynthetic process(GO:0006177) |
| 0.0 |
0.2 |
GO:0034638 |
phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 |
0.6 |
GO:0006750 |
glutathione biosynthetic process(GO:0006750) |
| 0.0 |
0.1 |
GO:0036166 |
phenotypic switching(GO:0036166) |
| 0.0 |
0.6 |
GO:1903963 |
arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.0 |
0.3 |
GO:0034983 |
peptidyl-lysine deacetylation(GO:0034983) |
| 0.0 |
0.8 |
GO:0010738 |
regulation of protein kinase A signaling(GO:0010738) |
| 0.0 |
0.3 |
GO:0071578 |
zinc II ion transmembrane import(GO:0071578) |
| 0.0 |
0.2 |
GO:0048170 |
positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.0 |
0.2 |
GO:0061002 |
negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.0 |
0.3 |
GO:0048742 |
regulation of skeletal muscle fiber development(GO:0048742) |
| 0.0 |
0.6 |
GO:0016226 |
iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 |
0.3 |
GO:0042182 |
ketone catabolic process(GO:0042182) |
| 0.0 |
0.6 |
GO:0006829 |
zinc II ion transport(GO:0006829) |
| 0.0 |
1.1 |
GO:0048843 |
negative regulation of axon extension involved in axon guidance(GO:0048843) |
| 0.0 |
0.7 |
GO:0045722 |
positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 |
0.0 |
GO:0002295 |
T-helper cell lineage commitment(GO:0002295) interleukin-4-mediated signaling pathway(GO:0035771) |
| 0.0 |
0.1 |
GO:0051189 |
Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 |
0.1 |
GO:0070589 |
cell wall mannoprotein biosynthetic process(GO:0000032) mannoprotein metabolic process(GO:0006056) mannoprotein biosynthetic process(GO:0006057) cell wall glycoprotein biosynthetic process(GO:0031506) cell wall biogenesis(GO:0042546) cell wall macromolecule metabolic process(GO:0044036) cell wall macromolecule biosynthetic process(GO:0044038) chain elongation of O-linked mannose residue(GO:0044845) cellular component macromolecule biosynthetic process(GO:0070589) cell wall organization or biogenesis(GO:0071554) |
| 0.0 |
0.1 |
GO:0010986 |
positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.0 |
0.2 |
GO:1901250 |
regulation of lung goblet cell differentiation(GO:1901249) negative regulation of lung goblet cell differentiation(GO:1901250) |
| 0.0 |
0.8 |
GO:0090162 |
establishment of epithelial cell polarity(GO:0090162) |
| 0.0 |
0.3 |
GO:0043248 |
proteasome assembly(GO:0043248) |
| 0.0 |
0.9 |
GO:0006418 |
tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 |
0.1 |
GO:0016255 |
attachment of GPI anchor to protein(GO:0016255) |
| 0.0 |
2.5 |
GO:0010977 |
negative regulation of neuron projection development(GO:0010977) |
| 0.0 |
0.6 |
GO:0034314 |
Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 |
0.1 |
GO:0051967 |
negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 |
0.3 |
GO:0099517 |
anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 |
0.3 |
GO:0048268 |
clathrin coat assembly(GO:0048268) |
| 0.0 |
0.4 |
GO:0008045 |
motor neuron axon guidance(GO:0008045) |
| 0.0 |
0.2 |
GO:0060712 |
spongiotrophoblast layer development(GO:0060712) |
| 0.0 |
0.0 |
GO:1904431 |
positive regulation of t-circle formation(GO:1904431) |
| 0.0 |
0.2 |
GO:0000188 |
inactivation of MAPK activity(GO:0000188) |
| 0.0 |
1.1 |
GO:0051965 |
positive regulation of synapse assembly(GO:0051965) |
| 0.0 |
2.1 |
GO:0071805 |
potassium ion transmembrane transport(GO:0071805) |
| 0.0 |
0.5 |
GO:1902476 |
chloride transmembrane transport(GO:1902476) |
| 0.0 |
0.1 |
GO:0006620 |
posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 |
0.3 |
GO:0007212 |
dopamine receptor signaling pathway(GO:0007212) |
| 0.0 |
0.4 |
GO:0006110 |
regulation of glycolytic process(GO:0006110) |
| 0.0 |
0.2 |
GO:0045838 |
positive regulation of membrane potential(GO:0045838) |
| 0.0 |
0.4 |
GO:0000002 |
mitochondrial genome maintenance(GO:0000002) |
| 0.0 |
0.7 |
GO:0001676 |
long-chain fatty acid metabolic process(GO:0001676) |
| 0.0 |
0.2 |
GO:0006054 |
N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 |
0.2 |
GO:0048557 |
embryonic digestive tract morphogenesis(GO:0048557) |