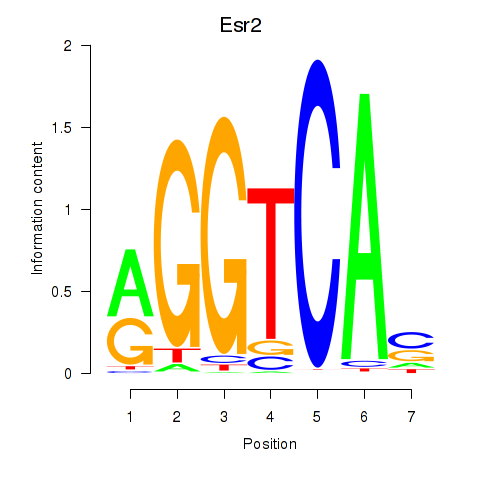
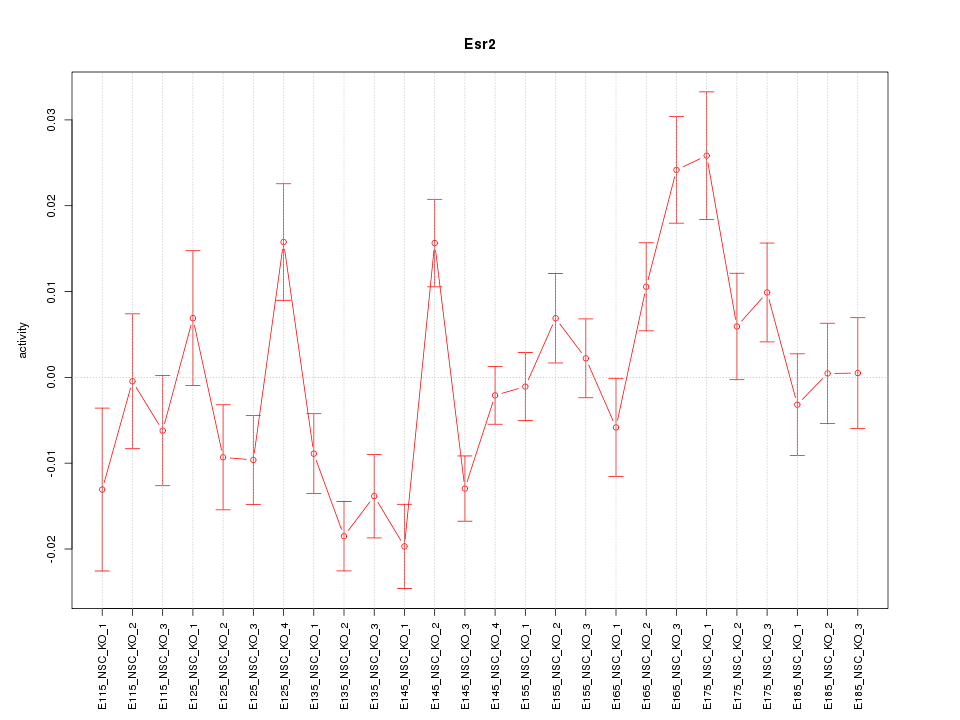
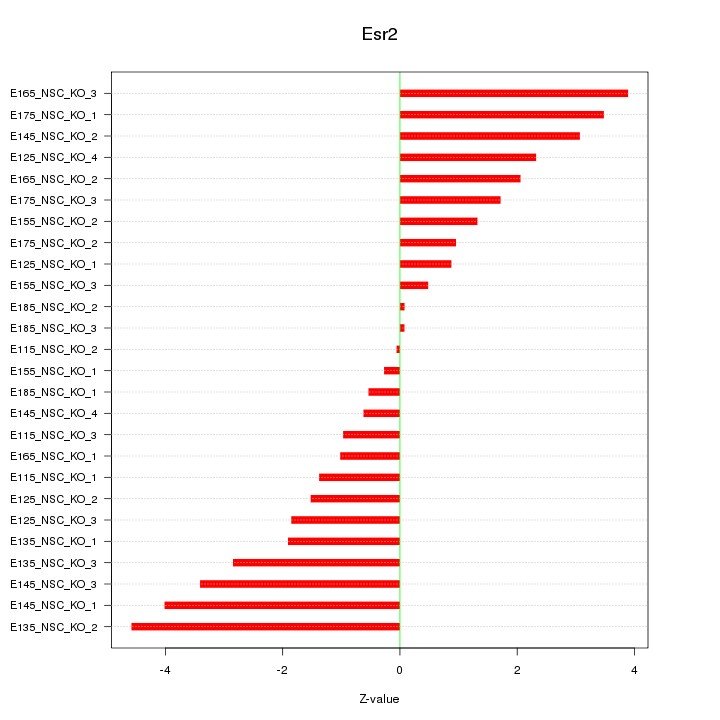
| 3.1 |
9.4 |
GO:0032430 |
positive regulation of phospholipase A2 activity(GO:0032430) |
| 2.0 |
6.1 |
GO:0071929 |
alpha-tubulin acetylation(GO:0071929) |
| 1.6 |
4.7 |
GO:2001139 |
negative regulation of postsynaptic membrane organization(GO:1901627) negative regulation of dendritic spine maintenance(GO:1902951) negative regulation of phospholipid efflux(GO:1902999) regulation of lipid transport across blood brain barrier(GO:1903000) negative regulation of lipid transport across blood brain barrier(GO:1903001) positive regulation of lipid transport across blood brain barrier(GO:1903002) negative regulation of phospholipid transport(GO:2001139) |
| 1.4 |
4.2 |
GO:0042560 |
10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 1.3 |
3.8 |
GO:0007521 |
muscle cell fate determination(GO:0007521) |
| 1.1 |
3.4 |
GO:0021933 |
radial glia guided migration of cerebellar granule cell(GO:0021933) |
| 1.1 |
3.3 |
GO:0043181 |
vacuolar sequestering(GO:0043181) |
| 1.1 |
3.3 |
GO:0032765 |
positive regulation of mast cell cytokine production(GO:0032765) |
| 1.0 |
5.0 |
GO:2000255 |
negative regulation of male germ cell proliferation(GO:2000255) |
| 1.0 |
1.0 |
GO:0048388 |
endosomal lumen acidification(GO:0048388) |
| 0.9 |
0.9 |
GO:0071726 |
response to diacyl bacterial lipopeptide(GO:0071724) cellular response to diacyl bacterial lipopeptide(GO:0071726) |
| 0.8 |
1.7 |
GO:0001803 |
type III hypersensitivity(GO:0001802) regulation of type III hypersensitivity(GO:0001803) positive regulation of type III hypersensitivity(GO:0001805) |
| 0.8 |
6.2 |
GO:0097460 |
ferrous iron import into cell(GO:0097460) |
| 0.7 |
2.2 |
GO:0060785 |
regulation of apoptosis involved in tissue homeostasis(GO:0060785) |
| 0.7 |
7.0 |
GO:0097421 |
liver regeneration(GO:0097421) |
| 0.7 |
2.1 |
GO:1900126 |
negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.7 |
4.2 |
GO:0006172 |
ADP biosynthetic process(GO:0006172) |
| 0.7 |
2.0 |
GO:0060478 |
acrosomal vesicle exocytosis(GO:0060478) |
| 0.7 |
2.7 |
GO:1902512 |
positive regulation of apoptotic DNA fragmentation(GO:1902512) positive regulation of DNA catabolic process(GO:1903626) |
| 0.7 |
2.7 |
GO:1900533 |
medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.7 |
4.0 |
GO:2000672 |
negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.6 |
2.5 |
GO:0014012 |
peripheral nervous system axon regeneration(GO:0014012) |
| 0.6 |
0.6 |
GO:0045763 |
negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 0.6 |
0.6 |
GO:2000346 |
negative regulation of hepatocyte proliferation(GO:2000346) positive regulation of integrin-mediated signaling pathway(GO:2001046) |
| 0.6 |
2.5 |
GO:0021564 |
glossopharyngeal nerve development(GO:0021563) vagus nerve development(GO:0021564) |
| 0.6 |
0.6 |
GO:0071374 |
cellular response to parathyroid hormone stimulus(GO:0071374) |
| 0.6 |
1.8 |
GO:1902358 |
sulfate transmembrane transport(GO:1902358) |
| 0.6 |
0.6 |
GO:1901252 |
regulation of intracellular transport of viral material(GO:1901252) |
| 0.6 |
1.8 |
GO:0030827 |
negative regulation of cGMP metabolic process(GO:0030824) negative regulation of cGMP biosynthetic process(GO:0030827) negative regulation of guanylate cyclase activity(GO:0031283) |
| 0.6 |
4.1 |
GO:0045794 |
negative regulation of cell volume(GO:0045794) |
| 0.6 |
2.3 |
GO:0001920 |
negative regulation of receptor recycling(GO:0001920) |
| 0.6 |
0.6 |
GO:1903181 |
regulation of dopamine biosynthetic process(GO:1903179) positive regulation of dopamine biosynthetic process(GO:1903181) |
| 0.6 |
7.4 |
GO:0043589 |
skin morphogenesis(GO:0043589) |
| 0.6 |
1.7 |
GO:0070327 |
thyroid hormone transport(GO:0070327) |
| 0.6 |
1.7 |
GO:1903795 |
regulation of inorganic anion transmembrane transport(GO:1903795) |
| 0.5 |
3.8 |
GO:0045650 |
negative regulation of macrophage differentiation(GO:0045650) |
| 0.5 |
2.2 |
GO:1901204 |
regulation of adrenergic receptor signaling pathway involved in heart process(GO:1901204) |
| 0.5 |
2.6 |
GO:0070459 |
prolactin secretion(GO:0070459) |
| 0.5 |
1.6 |
GO:0035435 |
phosphate ion transmembrane transport(GO:0035435) |
| 0.5 |
0.5 |
GO:0018208 |
peptidyl-proline modification(GO:0018208) |
| 0.5 |
1.5 |
GO:0017055 |
negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 0.5 |
1.0 |
GO:0090191 |
negative regulation of branching involved in ureteric bud morphogenesis(GO:0090191) |
| 0.5 |
3.0 |
GO:0006957 |
complement activation, alternative pathway(GO:0006957) |
| 0.5 |
1.5 |
GO:2000501 |
natural killer cell chemotaxis(GO:0035747) negative regulation of lymphocyte migration(GO:2000402) regulation of natural killer cell chemotaxis(GO:2000501) |
| 0.5 |
2.4 |
GO:0046016 |
regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) positive regulation of transcription by glucose(GO:0046016) |
| 0.5 |
3.3 |
GO:0061002 |
negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.5 |
1.4 |
GO:2001206 |
positive regulation of osteoclast development(GO:2001206) |
| 0.5 |
0.5 |
GO:0032489 |
regulation of Cdc42 protein signal transduction(GO:0032489) |
| 0.5 |
2.8 |
GO:0072318 |
clathrin coat disassembly(GO:0072318) |
| 0.5 |
1.4 |
GO:2000344 |
positive regulation of acrosome reaction(GO:2000344) |
| 0.5 |
5.1 |
GO:0097688 |
AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.5 |
2.8 |
GO:0030913 |
paranodal junction assembly(GO:0030913) |
| 0.5 |
1.4 |
GO:0007341 |
penetration of zona pellucida(GO:0007341) |
| 0.5 |
1.8 |
GO:0060050 |
positive regulation of protein glycosylation(GO:0060050) |
| 0.5 |
1.4 |
GO:0035441 |
cell migration involved in vasculogenesis(GO:0035441) |
| 0.4 |
4.0 |
GO:0097091 |
synaptic vesicle clustering(GO:0097091) |
| 0.4 |
0.9 |
GO:0036166 |
phenotypic switching(GO:0036166) |
| 0.4 |
1.7 |
GO:0051708 |
intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) cytosol to ER transport(GO:0046967) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.4 |
2.2 |
GO:0031915 |
positive regulation of synaptic plasticity(GO:0031915) |
| 0.4 |
3.0 |
GO:0033227 |
dsRNA transport(GO:0033227) |
| 0.4 |
1.7 |
GO:0010735 |
positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.4 |
1.7 |
GO:0016557 |
peroxisome membrane biogenesis(GO:0016557) |
| 0.4 |
4.3 |
GO:0036376 |
sodium ion export from cell(GO:0036376) |
| 0.4 |
1.3 |
GO:1900736 |
regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900736) |
| 0.4 |
1.2 |
GO:0071492 |
cellular response to UV-A(GO:0071492) |
| 0.4 |
1.2 |
GO:1904457 |
positive regulation of neuronal action potential(GO:1904457) |
| 0.4 |
1.2 |
GO:0042196 |
dichloromethane metabolic process(GO:0018900) chlorinated hydrocarbon metabolic process(GO:0042196) halogenated hydrocarbon metabolic process(GO:0042197) |
| 0.4 |
1.6 |
GO:0042167 |
heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.4 |
1.2 |
GO:0007210 |
serotonin receptor signaling pathway(GO:0007210) |
| 0.4 |
1.6 |
GO:0034975 |
protein folding in endoplasmic reticulum(GO:0034975) |
| 0.4 |
0.8 |
GO:0032417 |
positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 0.4 |
2.4 |
GO:0014053 |
negative regulation of gamma-aminobutyric acid secretion(GO:0014053) |
| 0.4 |
1.6 |
GO:0060448 |
dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.4 |
2.3 |
GO:0001955 |
blood vessel maturation(GO:0001955) |
| 0.4 |
1.9 |
GO:0043314 |
negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.4 |
1.9 |
GO:0030259 |
lipid glycosylation(GO:0030259) |
| 0.4 |
1.2 |
GO:0014738 |
regulation of muscle hyperplasia(GO:0014738) muscle hyperplasia(GO:0014900) |
| 0.4 |
1.1 |
GO:1900365 |
positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.4 |
0.8 |
GO:0032713 |
negative regulation of interleukin-4 production(GO:0032713) |
| 0.4 |
0.4 |
GO:1901897 |
regulation of relaxation of cardiac muscle(GO:1901897) |
| 0.4 |
1.9 |
GO:0018364 |
peptidyl-glutamine methylation(GO:0018364) |
| 0.4 |
0.7 |
GO:2000551 |
regulation of T-helper 2 cell cytokine production(GO:2000551) positive regulation of T-helper 2 cell cytokine production(GO:2000553) |
| 0.4 |
1.1 |
GO:0010871 |
negative regulation of receptor biosynthetic process(GO:0010871) |
| 0.4 |
1.1 |
GO:0046341 |
CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.4 |
1.5 |
GO:0051541 |
elastin metabolic process(GO:0051541) |
| 0.4 |
0.4 |
GO:0006851 |
mitochondrial calcium ion transport(GO:0006851) |
| 0.4 |
3.3 |
GO:0060770 |
negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.4 |
1.8 |
GO:0010359 |
regulation of anion channel activity(GO:0010359) |
| 0.4 |
1.8 |
GO:0002024 |
diet induced thermogenesis(GO:0002024) adaptive thermogenesis(GO:1990845) |
| 0.4 |
1.4 |
GO:0070358 |
actin polymerization-dependent cell motility(GO:0070358) |
| 0.4 |
4.7 |
GO:0043252 |
sodium-independent organic anion transport(GO:0043252) |
| 0.4 |
0.7 |
GO:0071677 |
positive regulation of mononuclear cell migration(GO:0071677) |
| 0.4 |
1.1 |
GO:0072531 |
pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.4 |
1.8 |
GO:0045347 |
negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.4 |
0.4 |
GO:2000830 |
vacuolar phosphate transport(GO:0007037) vitamin D3 metabolic process(GO:0070640) positive regulation of parathyroid hormone secretion(GO:2000830) |
| 0.4 |
0.4 |
GO:0034287 |
detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.4 |
1.1 |
GO:0071603 |
endothelial cell-cell adhesion(GO:0071603) |
| 0.3 |
1.7 |
GO:2001025 |
positive regulation of response to drug(GO:2001025) |
| 0.3 |
3.1 |
GO:0019800 |
peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.3 |
1.4 |
GO:0060265 |
positive regulation of respiratory burst involved in inflammatory response(GO:0060265) |
| 0.3 |
1.0 |
GO:0072008 |
glomerular mesangial cell differentiation(GO:0072008) glomerular mesangial cell development(GO:0072144) |
| 0.3 |
1.0 |
GO:0046959 |
habituation(GO:0046959) |
| 0.3 |
0.7 |
GO:0003105 |
negative regulation of glomerular filtration(GO:0003105) |
| 0.3 |
2.7 |
GO:0031666 |
positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.3 |
1.3 |
GO:0006432 |
phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.3 |
8.4 |
GO:0032011 |
ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.3 |
0.3 |
GO:0036115 |
fatty-acyl-CoA catabolic process(GO:0036115) |
| 0.3 |
2.0 |
GO:0006123 |
mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.3 |
1.0 |
GO:0048611 |
ectodermal digestive tract development(GO:0007439) embryonic ectodermal digestive tract development(GO:0048611) |
| 0.3 |
1.6 |
GO:0051012 |
microtubule sliding(GO:0051012) negative regulation of nonmotile primary cilium assembly(GO:1902856) |
| 0.3 |
1.6 |
GO:0032902 |
nerve growth factor production(GO:0032902) |
| 0.3 |
0.6 |
GO:0061588 |
calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylserine scrambling(GO:0061589) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.3 |
1.0 |
GO:0010807 |
regulation of synaptic vesicle priming(GO:0010807) |
| 0.3 |
1.3 |
GO:0072048 |
pattern specification involved in kidney development(GO:0061004) renal system pattern specification(GO:0072048) |
| 0.3 |
1.6 |
GO:0051365 |
cellular response to potassium ion starvation(GO:0051365) |
| 0.3 |
0.3 |
GO:1902445 |
regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.3 |
0.9 |
GO:0048818 |
positive regulation of hair follicle maturation(GO:0048818) positive regulation of catagen(GO:0051795) |
| 0.3 |
0.9 |
GO:0046168 |
glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.3 |
0.9 |
GO:0001193 |
maintenance of transcriptional fidelity during DNA-templated transcription elongation(GO:0001192) maintenance of transcriptional fidelity during DNA-templated transcription elongation from RNA polymerase II promoter(GO:0001193) |
| 0.3 |
1.9 |
GO:0034316 |
negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.3 |
1.2 |
GO:1904425 |
negative regulation of GTP binding(GO:1904425) |
| 0.3 |
0.9 |
GO:0039521 |
suppression by virus of host autophagy(GO:0039521) negative regulation of sphingolipid biosynthesis involved in cellular sphingolipid homeostasis(GO:0090157) |
| 0.3 |
1.6 |
GO:0015671 |
oxygen transport(GO:0015671) |
| 0.3 |
0.9 |
GO:0048294 |
negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.3 |
0.6 |
GO:1902995 |
regulation of phospholipid efflux(GO:1902994) positive regulation of phospholipid efflux(GO:1902995) |
| 0.3 |
0.9 |
GO:0035799 |
ureter maturation(GO:0035799) |
| 0.3 |
3.9 |
GO:0033623 |
regulation of integrin activation(GO:0033623) |
| 0.3 |
0.6 |
GO:0045764 |
positive regulation of cellular amino acid metabolic process(GO:0045764) |
| 0.3 |
5.0 |
GO:0042541 |
hemoglobin biosynthetic process(GO:0042541) |
| 0.3 |
0.6 |
GO:0006116 |
NADH oxidation(GO:0006116) |
| 0.3 |
0.9 |
GO:0045113 |
regulation of integrin biosynthetic process(GO:0045113) |
| 0.3 |
1.2 |
GO:0002606 |
positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.3 |
1.5 |
GO:0048069 |
eye pigmentation(GO:0048069) |
| 0.3 |
1.2 |
GO:0030397 |
membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.3 |
1.2 |
GO:0070318 |
positive regulation of G0 to G1 transition(GO:0070318) |
| 0.3 |
0.3 |
GO:0048289 |
isotype switching to IgE isotypes(GO:0048289) regulation of isotype switching to IgE isotypes(GO:0048293) |
| 0.3 |
0.9 |
GO:0032226 |
positive regulation of synaptic transmission, dopaminergic(GO:0032226) |
| 0.3 |
3.7 |
GO:0021683 |
cerebellar granular layer morphogenesis(GO:0021683) |
| 0.3 |
1.1 |
GO:0031953 |
negative regulation of protein autophosphorylation(GO:0031953) |
| 0.3 |
1.1 |
GO:0010756 |
positive regulation of plasminogen activation(GO:0010756) |
| 0.3 |
1.7 |
GO:1903071 |
positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.3 |
1.4 |
GO:2000574 |
regulation of microtubule motor activity(GO:2000574) |
| 0.3 |
1.1 |
GO:0003430 |
growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.3 |
0.6 |
GO:0051151 |
negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.3 |
0.8 |
GO:1990035 |
calcium ion import into cell(GO:1990035) |
| 0.3 |
0.8 |
GO:0032513 |
negative regulation of protein phosphatase type 2B activity(GO:0032513) |
| 0.3 |
1.7 |
GO:1901029 |
negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.3 |
4.2 |
GO:0045792 |
negative regulation of cell size(GO:0045792) |
| 0.3 |
1.4 |
GO:0007197 |
adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.3 |
3.9 |
GO:0016559 |
peroxisome fission(GO:0016559) |
| 0.3 |
0.8 |
GO:0035590 |
purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.3 |
5.3 |
GO:0045956 |
positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.3 |
0.8 |
GO:2000813 |
negative regulation of barbed-end actin filament capping(GO:2000813) |
| 0.3 |
3.9 |
GO:0031340 |
positive regulation of vesicle fusion(GO:0031340) |
| 0.3 |
0.8 |
GO:0071895 |
odontoblast differentiation(GO:0071895) |
| 0.3 |
1.4 |
GO:0001844 |
protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:0001844) |
| 0.3 |
1.1 |
GO:0097167 |
circadian regulation of translation(GO:0097167) |
| 0.3 |
0.8 |
GO:0008626 |
granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.3 |
1.1 |
GO:0030223 |
neutrophil differentiation(GO:0030223) |
| 0.3 |
0.8 |
GO:0032241 |
positive regulation of nucleobase-containing compound transport(GO:0032241) regulation of nucleoside transport(GO:0032242) |
| 0.3 |
1.6 |
GO:0015015 |
heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.3 |
3.2 |
GO:0045161 |
neuronal ion channel clustering(GO:0045161) |
| 0.3 |
0.5 |
GO:0006590 |
thyroid hormone generation(GO:0006590) |
| 0.3 |
1.1 |
GO:0030836 |
positive regulation of actin filament depolymerization(GO:0030836) |
| 0.3 |
2.4 |
GO:0060294 |
cilium movement involved in cell motility(GO:0060294) |
| 0.3 |
1.6 |
GO:0036233 |
glycine import(GO:0036233) |
| 0.3 |
3.2 |
GO:0006883 |
cellular sodium ion homeostasis(GO:0006883) |
| 0.3 |
3.4 |
GO:0015812 |
gamma-aminobutyric acid transport(GO:0015812) |
| 0.3 |
2.1 |
GO:0047496 |
vesicle transport along microtubule(GO:0047496) |
| 0.3 |
0.8 |
GO:0039703 |
viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.3 |
1.8 |
GO:0051036 |
regulation of endosome size(GO:0051036) |
| 0.3 |
2.1 |
GO:1902947 |
regulation of tau-protein kinase activity(GO:1902947) |
| 0.3 |
1.0 |
GO:0043461 |
proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.3 |
1.0 |
GO:0055095 |
lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) |
| 0.3 |
1.3 |
GO:1905098 |
negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.3 |
1.8 |
GO:1903423 |
positive regulation of synaptic vesicle transport(GO:1902805) positive regulation of synaptic vesicle recycling(GO:1903423) |
| 0.3 |
0.8 |
GO:1903525 |
regulation of membrane tubulation(GO:1903525) |
| 0.3 |
3.3 |
GO:1901898 |
negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.3 |
0.8 |
GO:0030854 |
positive regulation of granulocyte differentiation(GO:0030854) |
| 0.3 |
0.8 |
GO:0070317 |
negative regulation of G0 to G1 transition(GO:0070317) |
| 0.2 |
1.2 |
GO:1902683 |
regulation of receptor localization to synapse(GO:1902683) |
| 0.2 |
0.5 |
GO:1990253 |
cellular response to leucine starvation(GO:1990253) |
| 0.2 |
2.0 |
GO:0061000 |
negative regulation of dendritic spine development(GO:0061000) |
| 0.2 |
0.7 |
GO:0060083 |
smooth muscle contraction involved in micturition(GO:0060083) |
| 0.2 |
1.0 |
GO:0072235 |
distal convoluted tubule development(GO:0072025) DCT cell differentiation(GO:0072069) metanephric distal convoluted tubule development(GO:0072221) metanephric distal tubule development(GO:0072235) metanephric DCT cell differentiation(GO:0072240) |
| 0.2 |
1.0 |
GO:0097428 |
protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.2 |
1.0 |
GO:0097494 |
regulation of vesicle size(GO:0097494) |
| 0.2 |
0.7 |
GO:2001013 |
epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.2 |
0.5 |
GO:2000253 |
positive regulation of feeding behavior(GO:2000253) |
| 0.2 |
1.0 |
GO:0038128 |
ERBB2 signaling pathway(GO:0038128) |
| 0.2 |
1.0 |
GO:2000823 |
regulation of androgen receptor activity(GO:2000823) |
| 0.2 |
1.4 |
GO:0007258 |
JUN phosphorylation(GO:0007258) |
| 0.2 |
0.7 |
GO:1902310 |
positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.2 |
0.7 |
GO:0008228 |
opsonization(GO:0008228) |
| 0.2 |
0.7 |
GO:0009193 |
pyrimidine ribonucleoside diphosphate metabolic process(GO:0009193) UDP metabolic process(GO:0046048) |
| 0.2 |
2.1 |
GO:0051386 |
regulation of neurotrophin TRK receptor signaling pathway(GO:0051386) |
| 0.2 |
4.4 |
GO:0022401 |
desensitization of G-protein coupled receptor protein signaling pathway(GO:0002029) negative adaptation of signaling pathway(GO:0022401) |
| 0.2 |
0.7 |
GO:0019478 |
D-amino acid catabolic process(GO:0019478) |
| 0.2 |
0.5 |
GO:1901856 |
negative regulation of cellular respiration(GO:1901856) |
| 0.2 |
2.1 |
GO:0060136 |
embryonic process involved in female pregnancy(GO:0060136) |
| 0.2 |
0.7 |
GO:0051482 |
positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.2 |
1.9 |
GO:0032264 |
IMP salvage(GO:0032264) |
| 0.2 |
1.6 |
GO:0018344 |
protein geranylgeranylation(GO:0018344) |
| 0.2 |
0.5 |
GO:0001543 |
ovarian follicle rupture(GO:0001543) |
| 0.2 |
0.5 |
GO:0048170 |
positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.2 |
0.5 |
GO:0032700 |
negative regulation of interleukin-17 production(GO:0032700) |
| 0.2 |
9.4 |
GO:0048821 |
erythrocyte development(GO:0048821) |
| 0.2 |
1.1 |
GO:0019227 |
neuronal action potential propagation(GO:0019227) response to pyrethroid(GO:0046684) action potential propagation(GO:0098870) |
| 0.2 |
0.5 |
GO:0033700 |
phospholipid efflux(GO:0033700) |
| 0.2 |
0.7 |
GO:1904294 |
positive regulation of ERAD pathway(GO:1904294) |
| 0.2 |
0.5 |
GO:0072321 |
chaperone-mediated protein transport(GO:0072321) |
| 0.2 |
2.5 |
GO:0032482 |
Rab protein signal transduction(GO:0032482) |
| 0.2 |
0.4 |
GO:0003245 |
cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.2 |
0.7 |
GO:2001286 |
regulation of caveolin-mediated endocytosis(GO:2001286) |
| 0.2 |
0.7 |
GO:0032474 |
otolith morphogenesis(GO:0032474) |
| 0.2 |
3.3 |
GO:2000251 |
positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.2 |
3.3 |
GO:2000060 |
positive regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000060) |
| 0.2 |
0.7 |
GO:0006642 |
triglyceride mobilization(GO:0006642) |
| 0.2 |
0.9 |
GO:0060179 |
male mating behavior(GO:0060179) |
| 0.2 |
1.5 |
GO:0007520 |
myoblast fusion(GO:0007520) |
| 0.2 |
0.2 |
GO:2000277 |
positive regulation of oxidative phosphorylation uncoupler activity(GO:2000277) |
| 0.2 |
1.3 |
GO:0018149 |
peptide cross-linking(GO:0018149) |
| 0.2 |
9.5 |
GO:0048843 |
negative regulation of axon extension involved in axon guidance(GO:0048843) |
| 0.2 |
0.6 |
GO:0018343 |
protein farnesylation(GO:0018343) |
| 0.2 |
2.8 |
GO:0048172 |
regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.2 |
1.5 |
GO:0006102 |
isocitrate metabolic process(GO:0006102) |
| 0.2 |
0.2 |
GO:0070309 |
lens fiber cell morphogenesis(GO:0070309) |
| 0.2 |
0.6 |
GO:0036481 |
intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:0036481) |
| 0.2 |
0.6 |
GO:0001542 |
ovulation from ovarian follicle(GO:0001542) |
| 0.2 |
0.6 |
GO:0009298 |
GDP-mannose biosynthetic process(GO:0009298) |
| 0.2 |
1.5 |
GO:0021894 |
cerebral cortex GABAergic interneuron development(GO:0021894) |
| 0.2 |
1.0 |
GO:0021993 |
initiation of neural tube closure(GO:0021993) |
| 0.2 |
2.3 |
GO:0006120 |
mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.2 |
1.2 |
GO:1901096 |
regulation of autophagosome maturation(GO:1901096) |
| 0.2 |
0.4 |
GO:0014873 |
response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 0.2 |
1.8 |
GO:0006122 |
mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.2 |
0.6 |
GO:1904211 |
membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.2 |
0.6 |
GO:0045908 |
negative regulation of vasodilation(GO:0045908) |
| 0.2 |
0.4 |
GO:0035990 |
tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.2 |
0.6 |
GO:0045041 |
protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.2 |
1.0 |
GO:0046598 |
positive regulation of viral entry into host cell(GO:0046598) |
| 0.2 |
0.6 |
GO:0015014 |
heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.2 |
0.6 |
GO:0007158 |
neuron cell-cell adhesion(GO:0007158) |
| 0.2 |
0.4 |
GO:0051892 |
negative regulation of cardioblast differentiation(GO:0051892) |
| 0.2 |
0.6 |
GO:0048102 |
autophagic cell death(GO:0048102) |
| 0.2 |
0.8 |
GO:0071499 |
response to laminar fluid shear stress(GO:0034616) cellular response to laminar fluid shear stress(GO:0071499) |
| 0.2 |
0.8 |
GO:0070086 |
ubiquitin-dependent endocytosis(GO:0070086) |
| 0.2 |
1.2 |
GO:0007253 |
cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.2 |
1.4 |
GO:0016082 |
synaptic vesicle priming(GO:0016082) |
| 0.2 |
3.1 |
GO:0010614 |
negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.2 |
1.2 |
GO:0005513 |
detection of calcium ion(GO:0005513) |
| 0.2 |
0.6 |
GO:0010499 |
proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.2 |
2.5 |
GO:0034497 |
protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.2 |
1.7 |
GO:0071569 |
protein ufmylation(GO:0071569) |
| 0.2 |
0.6 |
GO:0045657 |
positive regulation of monocyte differentiation(GO:0045657) |
| 0.2 |
2.5 |
GO:0007214 |
gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.2 |
0.8 |
GO:0042535 |
positive regulation of tumor necrosis factor biosynthetic process(GO:0042535) |
| 0.2 |
0.6 |
GO:1904742 |
regulation of telomeric DNA binding(GO:1904742) |
| 0.2 |
1.7 |
GO:0090084 |
negative regulation of inclusion body assembly(GO:0090084) |
| 0.2 |
0.4 |
GO:0034140 |
granuloma formation(GO:0002432) negative regulation of toll-like receptor 3 signaling pathway(GO:0034140) |
| 0.2 |
1.5 |
GO:0090385 |
phagosome-lysosome fusion(GO:0090385) |
| 0.2 |
4.8 |
GO:0015991 |
energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.2 |
2.0 |
GO:0019243 |
methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.2 |
0.7 |
GO:1904177 |
regulation of adipose tissue development(GO:1904177) |
| 0.2 |
0.9 |
GO:0010991 |
negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.2 |
0.2 |
GO:0033034 |
positive regulation of myeloid cell apoptotic process(GO:0033034) |
| 0.2 |
2.0 |
GO:2000480 |
negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.2 |
0.9 |
GO:0010624 |
regulation of Schwann cell proliferation(GO:0010624) |
| 0.2 |
1.8 |
GO:0042989 |
sequestering of actin monomers(GO:0042989) |
| 0.2 |
0.7 |
GO:2000671 |
regulation of motor neuron apoptotic process(GO:2000671) |
| 0.2 |
1.1 |
GO:0001539 |
cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.2 |
0.9 |
GO:0051593 |
response to folic acid(GO:0051593) |
| 0.2 |
1.6 |
GO:0060081 |
membrane hyperpolarization(GO:0060081) |
| 0.2 |
5.7 |
GO:0061098 |
positive regulation of protein tyrosine kinase activity(GO:0061098) |
| 0.2 |
3.2 |
GO:0007616 |
long-term memory(GO:0007616) |
| 0.2 |
0.2 |
GO:0042726 |
flavin-containing compound metabolic process(GO:0042726) |
| 0.2 |
0.5 |
GO:0097309 |
cap1 mRNA methylation(GO:0097309) |
| 0.2 |
4.2 |
GO:1901381 |
positive regulation of potassium ion transmembrane transport(GO:1901381) |
| 0.2 |
0.7 |
GO:0035461 |
vitamin transmembrane transport(GO:0035461) |
| 0.2 |
2.2 |
GO:0050832 |
defense response to fungus(GO:0050832) |
| 0.2 |
0.9 |
GO:0071955 |
recycling endosome to Golgi transport(GO:0071955) |
| 0.2 |
0.7 |
GO:0035902 |
response to immobilization stress(GO:0035902) |
| 0.2 |
0.5 |
GO:0016256 |
N-glycan processing to lysosome(GO:0016256) |
| 0.2 |
2.4 |
GO:0009072 |
aromatic amino acid family metabolic process(GO:0009072) |
| 0.2 |
0.7 |
GO:0042985 |
negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.2 |
0.3 |
GO:0035793 |
cell migration involved in kidney development(GO:0035787) cell migration involved in metanephros development(GO:0035788) metanephric mesenchymal cell migration(GO:0035789) positive regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:0035793) regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:1900238) regulation of metanephric mesenchymal cell migration(GO:2000589) positive regulation of metanephric mesenchymal cell migration(GO:2000591) |
| 0.2 |
0.7 |
GO:1990743 |
protein sialylation(GO:1990743) |
| 0.2 |
0.5 |
GO:1901727 |
positive regulation of deacetylase activity(GO:0090045) positive regulation of histone deacetylase activity(GO:1901727) |
| 0.2 |
0.5 |
GO:0044034 |
negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.2 |
0.5 |
GO:0060267 |
positive regulation of respiratory burst(GO:0060267) |
| 0.2 |
1.4 |
GO:0050965 |
sensory perception of temperature stimulus(GO:0050951) detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.2 |
2.2 |
GO:0014898 |
muscle hypertrophy in response to stress(GO:0003299) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.2 |
0.7 |
GO:0010992 |
ubiquitin homeostasis(GO:0010992) |
| 0.2 |
3.2 |
GO:0048268 |
clathrin coat assembly(GO:0048268) |
| 0.2 |
1.7 |
GO:0098903 |
regulation of membrane repolarization during action potential(GO:0098903) |
| 0.2 |
0.3 |
GO:0070682 |
proteasome regulatory particle assembly(GO:0070682) |
| 0.2 |
0.8 |
GO:0015808 |
L-alanine transport(GO:0015808) |
| 0.2 |
3.3 |
GO:0014047 |
glutamate secretion(GO:0014047) |
| 0.2 |
1.8 |
GO:0033622 |
integrin activation(GO:0033622) |
| 0.2 |
1.5 |
GO:0090051 |
negative regulation of cell migration involved in sprouting angiogenesis(GO:0090051) |
| 0.2 |
1.6 |
GO:0070131 |
positive regulation of mitochondrial translation(GO:0070131) |
| 0.2 |
0.5 |
GO:0097623 |
potassium ion export across plasma membrane(GO:0097623) regulation of potassium ion export(GO:1902302) |
| 0.2 |
0.3 |
GO:1904154 |
positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.2 |
1.5 |
GO:0051968 |
positive regulation of synaptic transmission, glutamatergic(GO:0051968) |
| 0.2 |
0.6 |
GO:1901491 |
negative regulation of lymphangiogenesis(GO:1901491) |
| 0.2 |
1.0 |
GO:0036444 |
calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.2 |
0.5 |
GO:0019074 |
viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.2 |
0.6 |
GO:0032788 |
saturated monocarboxylic acid metabolic process(GO:0032788) unsaturated monocarboxylic acid metabolic process(GO:0032789) |
| 0.2 |
0.3 |
GO:0009051 |
pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.2 |
0.6 |
GO:0045903 |
positive regulation of translational fidelity(GO:0045903) |
| 0.2 |
0.3 |
GO:0015816 |
glycine transport(GO:0015816) |
| 0.2 |
0.3 |
GO:0051084 |
'de novo' protein folding(GO:0006458) 'de novo' posttranslational protein folding(GO:0051084) |
| 0.2 |
2.6 |
GO:0032060 |
bleb assembly(GO:0032060) |
| 0.2 |
0.3 |
GO:0051684 |
maintenance of Golgi location(GO:0051684) |
| 0.2 |
0.6 |
GO:0043985 |
histone H4-R3 methylation(GO:0043985) |
| 0.2 |
0.5 |
GO:0090274 |
positive regulation of somatostatin secretion(GO:0090274) |
| 0.2 |
0.5 |
GO:0039536 |
negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.2 |
0.5 |
GO:0090285 |
regulation of protein glycosylation in Golgi(GO:0090283) negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.2 |
2.7 |
GO:0001990 |
regulation of systemic arterial blood pressure by hormone(GO:0001990) |
| 0.2 |
3.0 |
GO:0007205 |
protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.2 |
0.5 |
GO:0044268 |
multicellular organismal protein metabolic process(GO:0044268) |
| 0.2 |
5.0 |
GO:0032456 |
endocytic recycling(GO:0032456) |
| 0.2 |
2.2 |
GO:0061462 |
protein localization to lysosome(GO:0061462) |
| 0.2 |
1.3 |
GO:2000194 |
regulation of female gonad development(GO:2000194) |
| 0.2 |
2.3 |
GO:1900746 |
regulation of vascular endothelial growth factor signaling pathway(GO:1900746) |
| 0.2 |
0.3 |
GO:1901550 |
regulation of endothelial cell development(GO:1901550) regulation of establishment of endothelial barrier(GO:1903140) |
| 0.2 |
0.6 |
GO:0006049 |
UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.2 |
0.5 |
GO:0006103 |
2-oxoglutarate metabolic process(GO:0006103) |
| 0.2 |
0.3 |
GO:2000508 |
regulation of dendritic cell chemotaxis(GO:2000508) positive regulation of dendritic cell chemotaxis(GO:2000510) |
| 0.2 |
1.2 |
GO:2000467 |
positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.2 |
2.5 |
GO:0021819 |
layer formation in cerebral cortex(GO:0021819) |
| 0.2 |
0.5 |
GO:2000598 |
regulation of cyclin catabolic process(GO:2000598) negative regulation of cyclin catabolic process(GO:2000599) |
| 0.2 |
0.8 |
GO:0006020 |
inositol metabolic process(GO:0006020) |
| 0.2 |
0.5 |
GO:0046322 |
negative regulation of fatty acid oxidation(GO:0046322) |
| 0.2 |
0.6 |
GO:1901475 |
pyruvate transmembrane transport(GO:1901475) |
| 0.2 |
0.6 |
GO:2000483 |
negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.2 |
1.4 |
GO:0034776 |
response to histamine(GO:0034776) cellular response to histamine(GO:0071420) |
| 0.2 |
1.1 |
GO:0045602 |
negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.1 |
0.9 |
GO:0032782 |
bile acid secretion(GO:0032782) |
| 0.1 |
0.6 |
GO:0006689 |
ganglioside catabolic process(GO:0006689) |
| 0.1 |
0.7 |
GO:0015803 |
branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.1 |
0.4 |
GO:0048011 |
neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.1 |
0.7 |
GO:0043312 |
neutrophil degranulation(GO:0043312) |
| 0.1 |
0.3 |
GO:0060744 |
thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.1 |
1.0 |
GO:0006729 |
tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.1 |
1.2 |
GO:0035608 |
protein deglutamylation(GO:0035608) |
| 0.1 |
0.7 |
GO:0031293 |
membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.1 |
1.0 |
GO:0045056 |
transcytosis(GO:0045056) |
| 0.1 |
0.6 |
GO:0046007 |
negative regulation of activated T cell proliferation(GO:0046007) |
| 0.1 |
3.3 |
GO:0070584 |
mitochondrion morphogenesis(GO:0070584) |
| 0.1 |
1.0 |
GO:0060134 |
prepulse inhibition(GO:0060134) |
| 0.1 |
0.4 |
GO:0006420 |
arginyl-tRNA aminoacylation(GO:0006420) |
| 0.1 |
0.9 |
GO:0045607 |
regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.1 |
0.6 |
GO:0021886 |
hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) |
| 0.1 |
0.6 |
GO:0051725 |
protein de-ADP-ribosylation(GO:0051725) |
| 0.1 |
0.3 |
GO:2001016 |
positive regulation of skeletal muscle cell differentiation(GO:2001016) |
| 0.1 |
0.6 |
GO:0019287 |
isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.1 |
0.3 |
GO:0007066 |
female meiosis sister chromatid cohesion(GO:0007066) |
| 0.1 |
1.0 |
GO:0016139 |
glycoside catabolic process(GO:0016139) |
| 0.1 |
0.4 |
GO:0015886 |
heme transport(GO:0015886) |
| 0.1 |
0.8 |
GO:0034983 |
peptidyl-lysine deacetylation(GO:0034983) |
| 0.1 |
1.1 |
GO:0071260 |
cellular response to mechanical stimulus(GO:0071260) |
| 0.1 |
0.4 |
GO:0015844 |
monoamine transport(GO:0015844) |
| 0.1 |
1.2 |
GO:0002021 |
response to dietary excess(GO:0002021) |
| 0.1 |
0.5 |
GO:0019042 |
viral latency(GO:0019042) |
| 0.1 |
0.5 |
GO:2000507 |
positive regulation of energy homeostasis(GO:2000507) |
| 0.1 |
0.3 |
GO:2000409 |
positive regulation of T cell extravasation(GO:2000409) |
| 0.1 |
0.4 |
GO:0003096 |
renal sodium ion transport(GO:0003096) |
| 0.1 |
0.1 |
GO:0021562 |
vestibulocochlear nerve development(GO:0021562) |
| 0.1 |
0.8 |
GO:0046627 |
negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.1 |
0.8 |
GO:0006452 |
translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.1 |
0.3 |
GO:0051014 |
actin filament severing(GO:0051014) |
| 0.1 |
0.4 |
GO:0016561 |
protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.1 |
0.5 |
GO:0072675 |
osteoclast fusion(GO:0072675) |
| 0.1 |
1.4 |
GO:0045974 |
miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.1 |
0.4 |
GO:0060800 |
regulation of cell differentiation involved in embryonic placenta development(GO:0060800) Notch signaling involved in heart development(GO:0061314) |
| 0.1 |
0.3 |
GO:1903774 |
positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.1 |
0.1 |
GO:0035910 |
ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 0.1 |
0.9 |
GO:0060287 |
epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.1 |
0.9 |
GO:0015862 |
uridine transport(GO:0015862) |
| 0.1 |
0.6 |
GO:0008354 |
germ cell migration(GO:0008354) |
| 0.1 |
0.4 |
GO:0042732 |
D-xylose metabolic process(GO:0042732) |
| 0.1 |
0.4 |
GO:0034551 |
respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.1 |
2.2 |
GO:0099517 |
anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.1 |
0.8 |
GO:0007342 |
fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.1 |
0.9 |
GO:0006290 |
pyrimidine dimer repair(GO:0006290) |
| 0.1 |
0.6 |
GO:0019236 |
response to pheromone(GO:0019236) |
| 0.1 |
1.6 |
GO:1901032 |
negative regulation of response to reactive oxygen species(GO:1901032) negative regulation of hydrogen peroxide-induced cell death(GO:1903206) |
| 0.1 |
0.9 |
GO:0030382 |
sperm mitochondrion organization(GO:0030382) |
| 0.1 |
0.4 |
GO:0060080 |
inhibitory postsynaptic potential(GO:0060080) |
| 0.1 |
0.7 |
GO:0061635 |
regulation of protein complex stability(GO:0061635) |
| 0.1 |
0.4 |
GO:2000774 |
positive regulation of cellular senescence(GO:2000774) |
| 0.1 |
1.7 |
GO:0031115 |
negative regulation of microtubule polymerization(GO:0031115) |
| 0.1 |
0.5 |
GO:0090315 |
negative regulation of protein targeting to membrane(GO:0090315) |
| 0.1 |
1.4 |
GO:0002115 |
store-operated calcium entry(GO:0002115) |
| 0.1 |
0.4 |
GO:1903977 |
positive regulation of Schwann cell migration(GO:1900149) positive regulation of glial cell migration(GO:1903977) |
| 0.1 |
0.1 |
GO:0009169 |
purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.1 |
0.4 |
GO:0046032 |
ADP catabolic process(GO:0046032) |
| 0.1 |
0.2 |
GO:0070665 |
positive regulation of leukocyte proliferation(GO:0070665) |
| 0.1 |
0.4 |
GO:1990414 |
replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.1 |
4.0 |
GO:0034605 |
cellular response to heat(GO:0034605) |
| 0.1 |
0.6 |
GO:0006659 |
phosphatidylserine biosynthetic process(GO:0006659) |
| 0.1 |
0.4 |
GO:0010872 |
regulation of cholesterol esterification(GO:0010872) |
| 0.1 |
0.8 |
GO:0051152 |
positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.1 |
1.1 |
GO:0042407 |
cristae formation(GO:0042407) |
| 0.1 |
0.5 |
GO:0007128 |
meiotic prophase I(GO:0007128) prophase(GO:0051324) meiotic cell cycle phase(GO:0098762) meiosis I cell cycle phase(GO:0098764) |
| 0.1 |
0.2 |
GO:0045053 |
protein retention in Golgi apparatus(GO:0045053) |
| 0.1 |
0.3 |
GO:0033603 |
positive regulation of dopamine secretion(GO:0033603) |
| 0.1 |
0.2 |
GO:0032105 |
negative regulation of response to extracellular stimulus(GO:0032105) negative regulation of response to nutrient levels(GO:0032108) |
| 0.1 |
1.8 |
GO:0070831 |
basement membrane assembly(GO:0070831) |
| 0.1 |
0.1 |
GO:0048597 |
post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.1 |
0.7 |
GO:0030579 |
ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.1 |
0.3 |
GO:0032534 |
regulation of microvillus assembly(GO:0032534) |
| 0.1 |
1.3 |
GO:0032292 |
myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.1 |
0.5 |
GO:0000379 |
tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.1 |
1.7 |
GO:0032026 |
response to magnesium ion(GO:0032026) |
| 0.1 |
0.6 |
GO:0048305 |
immunoglobulin secretion(GO:0048305) |
| 0.1 |
1.0 |
GO:0009650 |
UV protection(GO:0009650) |
| 0.1 |
0.4 |
GO:0036465 |
synaptic vesicle recycling(GO:0036465) |
| 0.1 |
0.8 |
GO:0045200 |
establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.1 |
0.2 |
GO:0097368 |
establishment of Sertoli cell barrier(GO:0097368) |
| 0.1 |
0.1 |
GO:2000121 |
regulation of superoxide dismutase activity(GO:1901668) regulation of removal of superoxide radicals(GO:2000121) |
| 0.1 |
2.4 |
GO:0010737 |
protein kinase A signaling(GO:0010737) |
| 0.1 |
0.5 |
GO:0050910 |
detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.1 |
2.1 |
GO:0000028 |
ribosomal small subunit assembly(GO:0000028) |
| 0.1 |
2.1 |
GO:0015985 |
energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 |
0.4 |
GO:0050689 |
negative regulation of defense response to virus by host(GO:0050689) |
| 0.1 |
1.6 |
GO:0043496 |
regulation of protein homodimerization activity(GO:0043496) |
| 0.1 |
0.4 |
GO:0035116 |
embryonic hindlimb morphogenesis(GO:0035116) |
| 0.1 |
2.5 |
GO:1902476 |
chloride transmembrane transport(GO:1902476) |
| 0.1 |
1.7 |
GO:0032981 |
NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 |
0.4 |
GO:1902268 |
negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.1 |
1.4 |
GO:0051447 |
negative regulation of meiotic cell cycle(GO:0051447) |
| 0.1 |
0.2 |
GO:0042270 |
protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.1 |
0.5 |
GO:0048227 |
plasma membrane to endosome transport(GO:0048227) |
| 0.1 |
0.2 |
GO:0046671 |
negative regulation of retinal cell programmed cell death(GO:0046671) post-embryonic eye morphogenesis(GO:0048050) |
| 0.1 |
0.1 |
GO:1902953 |
positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) |
| 0.1 |
0.1 |
GO:0001788 |
antibody-dependent cellular cytotoxicity(GO:0001788) |
| 0.1 |
2.5 |
GO:0010043 |
response to zinc ion(GO:0010043) |
| 0.1 |
1.1 |
GO:0051560 |
mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.1 |
0.5 |
GO:0044130 |
negative regulation of growth of symbiont in host(GO:0044130) negative regulation of growth of symbiont involved in interaction with host(GO:0044146) |
| 0.1 |
0.3 |
GO:2000822 |
regulation of fear response(GO:1903365) regulation of behavioral fear response(GO:2000822) |
| 0.1 |
0.5 |
GO:0007021 |
tubulin complex assembly(GO:0007021) |
| 0.1 |
0.2 |
GO:1902774 |
late endosome to lysosome transport(GO:1902774) |
| 0.1 |
0.5 |
GO:0070236 |
negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.1 |
0.6 |
GO:0006013 |
mannose metabolic process(GO:0006013) |
| 0.1 |
0.3 |
GO:0051030 |
snRNA transport(GO:0051030) |
| 0.1 |
0.2 |
GO:1902219 |
maintenance of blood-brain barrier(GO:0035633) regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902218) negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.1 |
0.2 |
GO:0019852 |
L-ascorbic acid metabolic process(GO:0019852) |
| 0.1 |
1.2 |
GO:0051205 |
protein insertion into membrane(GO:0051205) |
| 0.1 |
0.3 |
GO:0035526 |
retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.1 |
2.4 |
GO:0030970 |
retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.1 |
1.7 |
GO:0019229 |
regulation of vasoconstriction(GO:0019229) |
| 0.1 |
0.2 |
GO:0061043 |
regulation of vascular wound healing(GO:0061043) |
| 0.1 |
1.3 |
GO:0009312 |
oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 |
0.7 |
GO:1901385 |
regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.1 |
0.7 |
GO:0043537 |
negative regulation of blood vessel endothelial cell migration(GO:0043537) |
| 0.1 |
0.6 |
GO:0035428 |
hexose transmembrane transport(GO:0035428) |
| 0.1 |
0.3 |
GO:0060761 |
negative regulation of response to cytokine stimulus(GO:0060761) |
| 0.1 |
0.5 |
GO:0051683 |
establishment of Golgi localization(GO:0051683) |
| 0.1 |
0.4 |
GO:0002093 |
auditory receptor cell morphogenesis(GO:0002093) |
| 0.1 |
0.3 |
GO:0000454 |
snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.1 |
0.5 |
GO:0002536 |
respiratory burst involved in inflammatory response(GO:0002536) respiratory burst involved in defense response(GO:0002679) |
| 0.1 |
0.8 |
GO:0018916 |
nitrobenzene metabolic process(GO:0018916) |
| 0.1 |
1.4 |
GO:0006744 |
ubiquinone biosynthetic process(GO:0006744) |
| 0.1 |
0.4 |
GO:2000544 |
cell chemotaxis to fibroblast growth factor(GO:0035766) endothelial cell chemotaxis to fibroblast growth factor(GO:0035768) regulation of cell chemotaxis to fibroblast growth factor(GO:1904847) positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000544) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.1 |
0.3 |
GO:0051835 |
positive regulation of synapse structural plasticity(GO:0051835) |
| 0.1 |
0.3 |
GO:0045950 |
telomeric loop formation(GO:0031627) negative regulation of mitotic recombination(GO:0045950) negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 0.1 |
0.5 |
GO:0021914 |
negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.1 |
0.3 |
GO:0002014 |
vasoconstriction of artery involved in ischemic response to lowering of systemic arterial blood pressure(GO:0002014) |
| 0.1 |
0.9 |
GO:0044804 |
nucleophagy(GO:0044804) |
| 0.1 |
1.6 |
GO:0071577 |
zinc II ion transmembrane transport(GO:0071577) |
| 0.1 |
0.3 |
GO:0010898 |
positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.1 |
0.5 |
GO:0048251 |
elastic fiber assembly(GO:0048251) |
| 0.1 |
0.2 |
GO:0072643 |
interferon-gamma secretion(GO:0072643) |
| 0.1 |
1.5 |
GO:0015701 |
bicarbonate transport(GO:0015701) |
| 0.1 |
0.5 |
GO:0010759 |
positive regulation of macrophage chemotaxis(GO:0010759) |
| 0.1 |
0.4 |
GO:0043987 |
histone H3-S10 phosphorylation(GO:0043987) |
| 0.1 |
0.4 |
GO:0023041 |
neuronal signal transduction(GO:0023041) |
| 0.1 |
1.8 |
GO:0006821 |
chloride transport(GO:0006821) |
| 0.1 |
0.5 |
GO:0050774 |
negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.1 |
0.4 |
GO:0072527 |
pyrimidine-containing compound metabolic process(GO:0072527) |
| 0.1 |
0.3 |
GO:0061743 |
motor learning(GO:0061743) |
| 0.1 |
0.3 |
GO:0032929 |
negative regulation of superoxide anion generation(GO:0032929) |
| 0.1 |
0.3 |
GO:0070124 |
mitochondrial translational initiation(GO:0070124) |
| 0.1 |
0.9 |
GO:0032411 |
positive regulation of transporter activity(GO:0032411) |
| 0.1 |
0.3 |
GO:0000189 |
MAPK import into nucleus(GO:0000189) |
| 0.1 |
0.2 |
GO:0023058 |
adaptation of signaling pathway(GO:0023058) |
| 0.1 |
0.3 |
GO:0040016 |
embryonic cleavage(GO:0040016) |
| 0.1 |
1.1 |
GO:0070842 |
aggresome assembly(GO:0070842) |
| 0.1 |
1.0 |
GO:0045838 |
positive regulation of membrane potential(GO:0045838) |
| 0.1 |
0.3 |
GO:0015695 |
organic cation transport(GO:0015695) |
| 0.1 |
0.6 |
GO:0006685 |
sphingomyelin catabolic process(GO:0006685) |
| 0.1 |
2.0 |
GO:0009408 |
response to heat(GO:0009408) |
| 0.1 |
0.8 |
GO:0051092 |
positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.1 |
0.8 |
GO:0006532 |
aspartate biosynthetic process(GO:0006532) |
| 0.1 |
0.2 |
GO:0001845 |
phagolysosome assembly(GO:0001845) |
| 0.1 |
0.2 |
GO:0071763 |
nuclear membrane organization(GO:0071763) |
| 0.1 |
1.1 |
GO:0032469 |
endoplasmic reticulum calcium ion homeostasis(GO:0032469) |
| 0.1 |
0.1 |
GO:1902460 |
regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.1 |
0.2 |
GO:0042637 |
catagen(GO:0042637) |
| 0.1 |
0.6 |
GO:0060628 |
regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.1 |
1.1 |
GO:1900037 |
regulation of cellular response to hypoxia(GO:1900037) |
| 0.1 |
1.7 |
GO:0071353 |
cellular response to interleukin-4(GO:0071353) |
| 0.1 |
0.2 |
GO:0006435 |
threonyl-tRNA aminoacylation(GO:0006435) |
| 0.1 |
0.6 |
GO:0071257 |
cellular response to electrical stimulus(GO:0071257) |
| 0.1 |
0.2 |
GO:0034370 |
triglyceride-rich lipoprotein particle remodeling(GO:0034370) |
| 0.1 |
0.2 |
GO:0030643 |
cellular phosphate ion homeostasis(GO:0030643) phosphate ion homeostasis(GO:0055062) cellular divalent inorganic anion homeostasis(GO:0072501) cellular trivalent inorganic anion homeostasis(GO:0072502) divalent inorganic anion homeostasis(GO:0072505) trivalent inorganic anion homeostasis(GO:0072506) |
| 0.1 |
0.6 |
GO:0035646 |
endosome to melanosome transport(GO:0035646) endosome to pigment granule transport(GO:0043485) pigment granule maturation(GO:0048757) |
| 0.1 |
2.6 |
GO:0072583 |
clathrin-mediated endocytosis(GO:0072583) |
| 0.1 |
0.1 |
GO:0050665 |
hydrogen peroxide biosynthetic process(GO:0050665) |
| 0.1 |
0.4 |
GO:0046208 |
spermine catabolic process(GO:0046208) |
| 0.1 |
0.8 |
GO:1901223 |
negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.1 |
0.3 |
GO:0030262 |
cellular component disassembly involved in execution phase of apoptosis(GO:0006921) apoptotic nuclear changes(GO:0030262) |
| 0.1 |
0.4 |
GO:0051694 |
pointed-end actin filament capping(GO:0051694) |
| 0.1 |
0.9 |
GO:0006465 |
signal peptide processing(GO:0006465) |
| 0.1 |
0.3 |
GO:0046456 |
icosanoid biosynthetic process(GO:0046456) fatty acid derivative biosynthetic process(GO:1901570) |
| 0.1 |
0.5 |
GO:0090129 |
positive regulation of synapse maturation(GO:0090129) |
| 0.1 |
0.6 |
GO:0006878 |
cellular copper ion homeostasis(GO:0006878) |
| 0.1 |
0.3 |
GO:1904587 |
glycoprotein ERAD pathway(GO:0097466) response to glycoprotein(GO:1904587) |
| 0.1 |
0.4 |
GO:0043535 |
regulation of blood vessel endothelial cell migration(GO:0043535) |
| 0.1 |
0.2 |
GO:0050904 |
diapedesis(GO:0050904) |
| 0.1 |
0.2 |
GO:0042364 |
water-soluble vitamin biosynthetic process(GO:0042364) |
| 0.1 |
0.8 |
GO:0060074 |
synapse maturation(GO:0060074) |
| 0.1 |
0.4 |
GO:0061086 |
negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.1 |
1.2 |
GO:0016226 |
iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.1 |
0.4 |
GO:0043388 |
positive regulation of DNA binding(GO:0043388) |
| 0.1 |
0.4 |
GO:0035025 |
positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.1 |
0.2 |
GO:0048681 |
negative regulation of axon regeneration(GO:0048681) |
| 0.1 |
0.5 |
GO:0035814 |
negative regulation of renal sodium excretion(GO:0035814) |
| 0.1 |
3.2 |
GO:0031338 |
regulation of vesicle fusion(GO:0031338) |
| 0.1 |
0.2 |
GO:0006434 |
seryl-tRNA aminoacylation(GO:0006434) |
| 0.1 |
0.2 |
GO:0032687 |
negative regulation of interferon-alpha production(GO:0032687) |
| 0.1 |
0.7 |
GO:0030238 |
male sex determination(GO:0030238) |
| 0.1 |
0.4 |
GO:0006646 |
phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 |
0.5 |
GO:0045672 |
positive regulation of osteoclast differentiation(GO:0045672) |
| 0.1 |
0.6 |
GO:1990126 |
retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.1 |
0.4 |
GO:0006857 |
oligopeptide transport(GO:0006857) |
| 0.1 |
0.4 |
GO:1901339 |
regulation of store-operated calcium channel activity(GO:1901339) |
| 0.1 |
0.1 |
GO:0033364 |
mast cell secretory granule organization(GO:0033364) |
| 0.1 |
2.9 |
GO:0010508 |
positive regulation of autophagy(GO:0010508) |
| 0.1 |
0.2 |
GO:0070589 |
cell wall mannoprotein biosynthetic process(GO:0000032) mannoprotein metabolic process(GO:0006056) mannoprotein biosynthetic process(GO:0006057) cell wall glycoprotein biosynthetic process(GO:0031506) cell wall biogenesis(GO:0042546) cell wall macromolecule metabolic process(GO:0044036) cell wall macromolecule biosynthetic process(GO:0044038) chain elongation of O-linked mannose residue(GO:0044845) cellular component macromolecule biosynthetic process(GO:0070589) cell wall organization or biogenesis(GO:0071554) |
| 0.1 |
0.3 |
GO:1901030 |
positive regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901030) |
| 0.1 |
0.4 |
GO:0051481 |
negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.1 |
0.4 |
GO:0006488 |
dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.1 |
0.5 |
GO:0007413 |
axonal fasciculation(GO:0007413) |
| 0.1 |
0.1 |
GO:2000418 |
positive regulation of eosinophil migration(GO:2000418) |
| 0.1 |
2.6 |
GO:0006890 |
retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.1 |
0.3 |
GO:0007585 |
respiratory gaseous exchange(GO:0007585) |
| 0.1 |
0.4 |
GO:0033601 |
positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.1 |
0.9 |
GO:0050995 |
negative regulation of lipid catabolic process(GO:0050995) |
| 0.1 |
0.3 |
GO:0090164 |
asymmetric Golgi ribbon formation(GO:0090164) |
| 0.1 |
0.2 |
GO:0001869 |
regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 0.1 |
0.3 |
GO:0090241 |
negative regulation of histone H4 acetylation(GO:0090241) |
| 0.1 |
0.9 |
GO:0007271 |
synaptic transmission, cholinergic(GO:0007271) |
| 0.1 |
0.7 |
GO:0015858 |
nucleoside transport(GO:0015858) |
| 0.1 |
0.2 |
GO:0043416 |
regulation of skeletal muscle tissue regeneration(GO:0043416) |
| 0.1 |
0.4 |
GO:0044068 |
modulation by symbiont of host cellular process(GO:0044068) |
| 0.1 |
0.1 |
GO:0071550 |
death-inducing signaling complex assembly(GO:0071550) |
| 0.1 |
0.9 |
GO:0030150 |
protein import into mitochondrial matrix(GO:0030150) |
| 0.1 |
0.3 |
GO:0019254 |
carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.1 |
1.0 |
GO:0046464 |
neutral lipid catabolic process(GO:0046461) acylglycerol catabolic process(GO:0046464) |
| 0.1 |
0.1 |
GO:0003376 |
sphingosine-1-phosphate signaling pathway(GO:0003376) sphingolipid mediated signaling pathway(GO:0090520) |
| 0.1 |
0.1 |
GO:0070257 |
positive regulation of mucus secretion(GO:0070257) |
| 0.1 |
0.5 |
GO:0010039 |
response to iron ion(GO:0010039) |
| 0.1 |
0.7 |
GO:0006047 |
UDP-N-acetylglucosamine metabolic process(GO:0006047) |
| 0.1 |
0.6 |
GO:1903020 |
positive regulation of glycoprotein metabolic process(GO:1903020) |
| 0.1 |
0.5 |
GO:0090043 |
tubulin deacetylation(GO:0090042) regulation of tubulin deacetylation(GO:0090043) |
| 0.1 |
0.8 |
GO:0051016 |
barbed-end actin filament capping(GO:0051016) |
| 0.1 |
1.4 |
GO:0034340 |
response to type I interferon(GO:0034340) |
| 0.1 |
1.0 |
GO:0005980 |
polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.1 |
0.3 |
GO:0035521 |
monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.1 |
0.9 |
GO:0035269 |
protein O-linked mannosylation(GO:0035269) |
| 0.1 |
0.9 |
GO:0046827 |
positive regulation of protein export from nucleus(GO:0046827) |
| 0.1 |
0.6 |
GO:0046040 |
IMP metabolic process(GO:0046040) |
| 0.1 |
0.4 |
GO:0046085 |
adenosine metabolic process(GO:0046085) |
| 0.1 |
0.2 |
GO:0000480 |
endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.1 |
0.4 |
GO:0043983 |
histone H4-K12 acetylation(GO:0043983) |
| 0.1 |
0.1 |
GO:1901896 |
positive regulation of calcium-transporting ATPase activity(GO:1901896) |
| 0.1 |
0.4 |
GO:0061092 |
regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.1 |
0.4 |
GO:2000653 |
regulation of genetic imprinting(GO:2000653) |
| 0.1 |
0.9 |
GO:0021516 |
dorsal spinal cord development(GO:0021516) |
| 0.1 |
0.2 |
GO:2000623 |
regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.1 |
0.2 |
GO:0001830 |
trophectodermal cell fate commitment(GO:0001830) |
| 0.1 |
0.3 |
GO:2000767 |
positive regulation of cytoplasmic translation(GO:2000767) |
| 0.1 |
0.3 |
GO:0042178 |
xenobiotic catabolic process(GO:0042178) |
| 0.1 |
0.2 |
GO:0014829 |
vascular smooth muscle contraction(GO:0014829) |
| 0.1 |
0.1 |
GO:0048211 |
Golgi vesicle docking(GO:0048211) |
| 0.1 |
0.2 |
GO:0034393 |
positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.1 |
1.1 |
GO:0001893 |
maternal placenta development(GO:0001893) |
| 0.1 |
1.7 |
GO:0034723 |
DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 |
0.4 |
GO:0070314 |
G1 to G0 transition(GO:0070314) |
| 0.1 |
0.2 |
GO:0002710 |
negative regulation of T cell mediated immunity(GO:0002710) |
| 0.1 |
0.5 |
GO:0033690 |
positive regulation of osteoblast proliferation(GO:0033690) |
| 0.1 |
0.8 |
GO:0006308 |
DNA catabolic process(GO:0006308) |
| 0.1 |
0.4 |
GO:0043457 |
regulation of cellular respiration(GO:0043457) |
| 0.1 |
1.3 |
GO:1904030 |
negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.1 |
0.3 |
GO:0032800 |
receptor biosynthetic process(GO:0032800) |
| 0.1 |
0.2 |
GO:0048169 |
regulation of long-term neuronal synaptic plasticity(GO:0048169) |
| 0.1 |
1.0 |
GO:0006904 |
vesicle docking involved in exocytosis(GO:0006904) |
| 0.1 |
0.4 |
GO:0001964 |
startle response(GO:0001964) |
| 0.1 |
0.5 |
GO:1900364 |
negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.1 |
0.2 |
GO:0015786 |
UDP-glucose transport(GO:0015786) |
| 0.1 |
1.2 |
GO:0046854 |
phosphatidylinositol phosphorylation(GO:0046854) |
| 0.1 |
0.8 |
GO:0002076 |
osteoblast development(GO:0002076) |
| 0.1 |
0.2 |
GO:0048311 |
mitochondrion distribution(GO:0048311) |
| 0.1 |
0.4 |
GO:0030828 |
positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.1 |
0.6 |
GO:0048280 |
vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.1 |
0.4 |
GO:0006390 |
transcription from mitochondrial promoter(GO:0006390) |
| 0.0 |
0.4 |
GO:0035881 |
amacrine cell differentiation(GO:0035881) |
| 0.0 |
0.2 |
GO:0055003 |
cardiac myofibril assembly(GO:0055003) |
| 0.0 |
0.2 |
GO:0045900 |
negative regulation of translational elongation(GO:0045900) |
| 0.0 |
0.2 |
GO:0009212 |
dTTP biosynthetic process(GO:0006235) deoxyribonucleoside triphosphate biosynthetic process(GO:0009202) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) dTTP metabolic process(GO:0046075) |
| 0.0 |
0.5 |
GO:0098792 |
xenophagy(GO:0098792) |
| 0.0 |
0.2 |
GO:0009437 |
carnitine metabolic process(GO:0009437) |
| 0.0 |
2.8 |
GO:0006487 |
protein N-linked glycosylation(GO:0006487) |
| 0.0 |
0.9 |
GO:0060765 |
regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.0 |
0.0 |
GO:0051957 |
positive regulation of amino acid transport(GO:0051957) |
| 0.0 |
0.1 |
GO:0032693 |
negative regulation of interleukin-10 production(GO:0032693) |
| 0.0 |
0.1 |
GO:0006669 |
sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.0 |
0.2 |
GO:0019673 |
GDP-mannose metabolic process(GO:0019673) |
| 0.0 |
0.8 |
GO:0002673 |
regulation of acute inflammatory response(GO:0002673) |
| 0.0 |
0.4 |
GO:0071941 |
nitrogen cycle metabolic process(GO:0071941) |
| 0.0 |
0.2 |
GO:0045026 |
plasma membrane fusion(GO:0045026) |
| 0.0 |
0.4 |
GO:0050901 |
leukocyte tethering or rolling(GO:0050901) |
| 0.0 |
0.5 |
GO:0070935 |
3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 |
0.3 |
GO:0018026 |
peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 |
0.3 |
GO:0035507 |
regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.0 |
0.6 |
GO:0021670 |
lateral ventricle development(GO:0021670) |
| 0.0 |
0.4 |
GO:0031167 |
rRNA methylation(GO:0031167) |
| 0.0 |
0.3 |
GO:0051193 |
regulation of cofactor metabolic process(GO:0051193) |
| 0.0 |
0.3 |
GO:0032525 |
somite rostral/caudal axis specification(GO:0032525) |
| 0.0 |
0.5 |
GO:0002098 |
tRNA wobble uridine modification(GO:0002098) |
| 0.0 |
0.1 |
GO:0002082 |
regulation of oxidative phosphorylation(GO:0002082) |
| 0.0 |
0.1 |
GO:1902170 |
cellular response to reactive nitrogen species(GO:1902170) |
| 0.0 |
0.2 |
GO:0007229 |
integrin-mediated signaling pathway(GO:0007229) |
| 0.0 |
0.3 |
GO:0044065 |
regulation of respiratory system process(GO:0044065) |
| 0.0 |
0.2 |
GO:0010387 |
COP9 signalosome assembly(GO:0010387) |
| 0.0 |
0.2 |
GO:0071494 |
cellular response to UV-C(GO:0071494) |
| 0.0 |
0.2 |
GO:1902993 |
positive regulation of beta-amyloid formation(GO:1902004) positive regulation of amyloid precursor protein catabolic process(GO:1902993) |
| 0.0 |
0.2 |
GO:0000395 |
mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 |
0.1 |
GO:2000765 |
regulation of cytoplasmic translation(GO:2000765) |
| 0.0 |
0.2 |
GO:0018410 |
C-terminal protein amino acid modification(GO:0018410) |
| 0.0 |
0.7 |
GO:0035036 |
binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) |
| 0.0 |
1.4 |
GO:0006414 |
translational elongation(GO:0006414) |
| 0.0 |
0.5 |
GO:0072520 |
seminiferous tubule development(GO:0072520) |
| 0.0 |
0.4 |
GO:0046051 |
UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.0 |
0.1 |
GO:0051189 |
Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 |
0.2 |
GO:0061051 |
positive regulation of cell growth involved in cardiac muscle cell development(GO:0061051) |
| 0.0 |
1.0 |
GO:0051148 |
negative regulation of muscle cell differentiation(GO:0051148) |
| 0.0 |
0.8 |
GO:0016525 |
negative regulation of angiogenesis(GO:0016525) |
| 0.0 |
0.2 |
GO:0006760 |
folic acid-containing compound metabolic process(GO:0006760) pteridine-containing compound metabolic process(GO:0042558) |
| 0.0 |
0.0 |
GO:0014043 |
negative regulation of neuron maturation(GO:0014043) |
| 0.0 |
0.8 |
GO:0032410 |
negative regulation of transporter activity(GO:0032410) |
| 0.0 |
0.2 |
GO:0097034 |
mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 |
0.2 |
GO:0006691 |
leukotriene metabolic process(GO:0006691) |
| 0.0 |
0.2 |
GO:0071985 |
multivesicular body sorting pathway(GO:0071985) |
| 0.0 |
0.2 |
GO:0061188 |
regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.0 |
1.2 |
GO:0006749 |
glutathione metabolic process(GO:0006749) |
| 0.0 |
0.1 |
GO:0044154 |
histone H3-K14 acetylation(GO:0044154) regulation of histone H3-K14 acetylation(GO:0071440) |
| 0.0 |
0.6 |
GO:0010107 |
potassium ion import(GO:0010107) |
| 0.0 |
1.1 |
GO:0010507 |
negative regulation of autophagy(GO:0010507) |
| 0.0 |
0.2 |
GO:0009448 |
gamma-aminobutyric acid metabolic process(GO:0009448) gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.0 |
0.4 |
GO:0035999 |
tetrahydrofolate interconversion(GO:0035999) |
| 0.0 |
0.3 |
GO:0015698 |
inorganic anion transport(GO:0015698) |
| 0.0 |
0.6 |
GO:0010971 |
positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) |
| 0.0 |
0.2 |
GO:0043949 |
regulation of cAMP-mediated signaling(GO:0043949) |
| 0.0 |
0.2 |
GO:0010818 |
T cell chemotaxis(GO:0010818) |
| 0.0 |
0.1 |
GO:0002634 |
regulation of germinal center formation(GO:0002634) |
| 0.0 |
0.4 |
GO:0032757 |
positive regulation of interleukin-8 production(GO:0032757) |
| 0.0 |
0.2 |
GO:0000305 |
response to oxygen radical(GO:0000305) |
| 0.0 |
0.0 |
GO:0006296 |
nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
| 0.0 |
0.1 |
GO:0046098 |
guanine metabolic process(GO:0046098) |
| 0.0 |
0.7 |
GO:0016079 |
synaptic vesicle exocytosis(GO:0016079) |
| 0.0 |
0.1 |
GO:0010566 |
regulation of ketone biosynthetic process(GO:0010566) |
| 0.0 |
0.1 |
GO:0033005 |
positive regulation of mast cell activation(GO:0033005) |
| 0.0 |
0.2 |
GO:0098532 |
histone H3-K27 trimethylation(GO:0098532) |
| 0.0 |
0.4 |
GO:0007097 |
nuclear migration(GO:0007097) |
| 0.0 |
1.1 |
GO:0032543 |
mitochondrial translation(GO:0032543) |
| 0.0 |
0.6 |
GO:0030032 |
lamellipodium assembly(GO:0030032) |
| 0.0 |
0.1 |
GO:0097011 |
cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) |
| 0.0 |
1.1 |
GO:0050775 |
positive regulation of dendrite morphogenesis(GO:0050775) |
| 0.0 |
0.3 |
GO:0044406 |
adhesion of symbiont to host(GO:0044406) |
| 0.0 |
0.1 |
GO:0003199 |
endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.0 |
0.4 |
GO:0006818 |
hydrogen transport(GO:0006818) |
| 0.0 |
0.4 |
GO:0006801 |
superoxide metabolic process(GO:0006801) |
| 0.0 |
0.2 |
GO:0050667 |
homocysteine metabolic process(GO:0050667) |
| 0.0 |
0.2 |
GO:0007416 |
synapse assembly(GO:0007416) |
| 0.0 |
0.2 |
GO:0030539 |
male genitalia development(GO:0030539) |
| 0.0 |
0.1 |
GO:0036112 |
medium-chain fatty-acyl-CoA metabolic process(GO:0036112) |
| 0.0 |
0.1 |
GO:1900017 |
positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.0 |
0.7 |
GO:0055013 |
cardiac muscle cell development(GO:0055013) |
| 0.0 |
1.2 |
GO:0042147 |
retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 |
0.2 |
GO:0006515 |
misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.0 |
0.2 |
GO:0006104 |
succinyl-CoA metabolic process(GO:0006104) |
| 0.0 |
0.1 |
GO:0007220 |
Notch receptor processing(GO:0007220) |
| 0.0 |
0.3 |
GO:1902235 |
regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902235) |
| 0.0 |
0.1 |
GO:0006361 |
transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.0 |
0.5 |
GO:0035335 |
peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 |
0.1 |
GO:0060481 |
lobar bronchus epithelium development(GO:0060481) |
| 0.0 |
0.1 |
GO:2000465 |
regulation of glycogen (starch) synthase activity(GO:2000465) |
| 0.0 |
2.2 |
GO:1903955 |
positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 |
0.3 |
GO:0034260 |
negative regulation of GTPase activity(GO:0034260) |
| 0.0 |
0.1 |
GO:0010724 |
regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 |
0.1 |
GO:0072592 |
oxygen metabolic process(GO:0072592) |
| 0.0 |
0.1 |
GO:0006573 |
valine metabolic process(GO:0006573) |
| 0.0 |
0.7 |
GO:0071806 |
intracellular protein transmembrane transport(GO:0065002) protein transmembrane transport(GO:0071806) |
| 0.0 |
0.5 |
GO:0032781 |
positive regulation of ATPase activity(GO:0032781) |
| 0.0 |
0.4 |
GO:0046325 |
negative regulation of glucose import(GO:0046325) |
| 0.0 |
0.2 |
GO:0051703 |
social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.0 |
0.2 |
GO:0014883 |
transition between fast and slow fiber(GO:0014883) |
| 0.0 |
0.1 |
GO:0006953 |
acute-phase response(GO:0006953) |
| 0.0 |
0.1 |
GO:0048711 |
positive regulation of astrocyte differentiation(GO:0048711) |
| 0.0 |
0.6 |
GO:0032728 |
positive regulation of interferon-beta production(GO:0032728) |
| 0.0 |
0.1 |
GO:2000152 |
regulation of protein K48-linked deubiquitination(GO:1903093) negative regulation of protein K48-linked deubiquitination(GO:1903094) regulation of ubiquitin-specific protease activity(GO:2000152) negative regulation of ubiquitin-specific protease activity(GO:2000157) |
| 0.0 |
0.1 |
GO:0046415 |
urate metabolic process(GO:0046415) |
| 0.0 |
0.2 |
GO:0051898 |
negative regulation of protein kinase B signaling(GO:0051898) |
| 0.0 |
0.1 |
GO:0035552 |
oxidative single-stranded DNA demethylation(GO:0035552) |
| 0.0 |
0.8 |
GO:0006376 |
mRNA splice site selection(GO:0006376) |
| 0.0 |
0.2 |
GO:0008298 |
intracellular mRNA localization(GO:0008298) |
| 0.0 |
0.1 |
GO:0034198 |
cellular response to amino acid starvation(GO:0034198) |
| 0.0 |
0.4 |
GO:0001945 |
lymph vessel development(GO:0001945) |
| 0.0 |
0.1 |
GO:1902108 |
regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902108) |
| 0.0 |
0.1 |
GO:0033152 |
immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 |
0.3 |
GO:0035249 |
synaptic transmission, glutamatergic(GO:0035249) |
| 0.0 |
1.6 |
GO:0007156 |
homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 |
0.3 |
GO:0046579 |
positive regulation of Ras protein signal transduction(GO:0046579) |
| 0.0 |
0.2 |
GO:0010388 |
cullin deneddylation(GO:0010388) |
| 0.0 |
0.2 |
GO:0071625 |
vocalization behavior(GO:0071625) |
| 0.0 |
0.2 |
GO:0006003 |
fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 |
0.1 |
GO:0050651 |
dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.0 |
0.3 |
GO:0090502 |
RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.0 |
0.1 |
GO:1902187 |
negative regulation of viral release from host cell(GO:1902187) |
| 0.0 |
0.4 |
GO:0016180 |
snRNA processing(GO:0016180) |
| 0.0 |
0.2 |
GO:0071542 |
dopaminergic neuron differentiation(GO:0071542) |
| 0.0 |
0.4 |
GO:0032008 |
positive regulation of TOR signaling(GO:0032008) |
| 0.0 |
0.2 |
GO:0070208 |
protein heterotrimerization(GO:0070208) |
| 0.0 |
0.4 |
GO:0019233 |
sensory perception of pain(GO:0019233) |
| 0.0 |
0.3 |
GO:0090503 |
RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.0 |
0.4 |
GO:0043153 |
photoperiodism(GO:0009648) entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 |
1.7 |
GO:0098780 |
mitophagy in response to mitochondrial depolarization(GO:0098779) response to mitochondrial depolarisation(GO:0098780) |
| 0.0 |
0.0 |
GO:1904431 |
positive regulation of t-circle formation(GO:1904431) |
| 0.0 |
0.2 |
GO:0070940 |
dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 |
0.1 |
GO:0006620 |
posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 |
0.1 |
GO:0030656 |
regulation of vitamin metabolic process(GO:0030656) |
| 0.0 |
0.1 |
GO:0006436 |
tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.0 |
0.3 |
GO:0048266 |
behavioral response to pain(GO:0048266) |
| 0.0 |
0.1 |
GO:1902035 |
regulation of hematopoietic stem cell proliferation(GO:1902033) positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.0 |
0.1 |
GO:0060632 |
regulation of microtubule-based movement(GO:0060632) |
| 0.0 |
0.2 |
GO:0050829 |
defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 |
0.1 |
GO:0008655 |
pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.0 |
0.0 |
GO:0036336 |
myeloid dendritic cell chemotaxis(GO:0002408) dendritic cell migration(GO:0036336) |
| 0.0 |
0.2 |
GO:0031641 |
regulation of myelination(GO:0031641) |
| 0.0 |
0.1 |
GO:0042755 |
eating behavior(GO:0042755) |
| 0.0 |
0.3 |
GO:1901998 |
toxin transport(GO:1901998) |
| 0.0 |
0.1 |
GO:0000715 |
nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.0 |
0.1 |
GO:0033147 |
negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 |
0.2 |
GO:0002227 |
innate immune response in mucosa(GO:0002227) |
| 0.0 |
0.1 |
GO:2000096 |
positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 |
0.1 |
GO:0042182 |
ketone catabolic process(GO:0042182) |
| 0.0 |
0.2 |
GO:0034244 |
negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 |
0.0 |
GO:0046339 |
diacylglycerol metabolic process(GO:0046339) |
| 0.0 |
0.1 |
GO:0035845 |
photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 |
0.1 |
GO:0018377 |
N-terminal protein myristoylation(GO:0006499) protein myristoylation(GO:0018377) |
| 0.0 |
0.1 |
GO:0071361 |
cellular response to ethanol(GO:0071361) |
| 0.0 |
0.1 |
GO:1903352 |
ornithine transport(GO:0015822) L-ornithine transmembrane transport(GO:1903352) |
| 0.0 |
0.1 |
GO:0006654 |
phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.0 |
0.2 |
GO:0030574 |
collagen catabolic process(GO:0030574) |
| 0.0 |
0.1 |
GO:1900027 |
regulation of ruffle assembly(GO:1900027) |
| 0.0 |
0.5 |
GO:1901216 |
positive regulation of neuron death(GO:1901216) |
| 0.0 |
0.2 |
GO:0009191 |
ribonucleoside diphosphate catabolic process(GO:0009191) |
| 0.0 |
0.2 |
GO:0019262 |
N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 |
0.3 |
GO:0014904 |
myotube cell development(GO:0014904) |
| 0.0 |
0.2 |
GO:0045921 |
positive regulation of exocytosis(GO:0045921) |
| 0.0 |
0.4 |
GO:0030968 |
endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 |
0.0 |
GO:0034310 |
primary alcohol catabolic process(GO:0034310) |
| 0.0 |
0.0 |
GO:0007256 |
activation of JNKK activity(GO:0007256) |
| 0.0 |
0.1 |
GO:0006144 |
purine nucleobase metabolic process(GO:0006144) |
| 0.0 |
0.1 |
GO:0038032 |
termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.0 |
0.0 |
GO:0032790 |
ribosome disassembly(GO:0032790) |
| 0.0 |
0.1 |
GO:0070301 |
cellular response to hydrogen peroxide(GO:0070301) |
| 0.0 |
0.5 |
GO:0043967 |
histone H4 acetylation(GO:0043967) |
| 0.0 |
0.1 |
GO:0032464 |
positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 |
0.0 |
GO:2000811 |
negative regulation of anoikis(GO:2000811) |
| 0.0 |
0.0 |
GO:0014742 |
positive regulation of cardiac muscle hypertrophy(GO:0010613) positive regulation of muscle hypertrophy(GO:0014742) |
| 0.0 |
0.2 |
GO:0003333 |
amino acid transmembrane transport(GO:0003333) |
| 0.0 |
0.2 |
GO:0031648 |
protein destabilization(GO:0031648) |
| 0.0 |
0.1 |
GO:0043518 |
negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.0 |
0.1 |
GO:0000103 |
sulfate assimilation(GO:0000103) |
| 0.0 |
0.1 |
GO:0035337 |
fatty-acyl-CoA metabolic process(GO:0035337) |