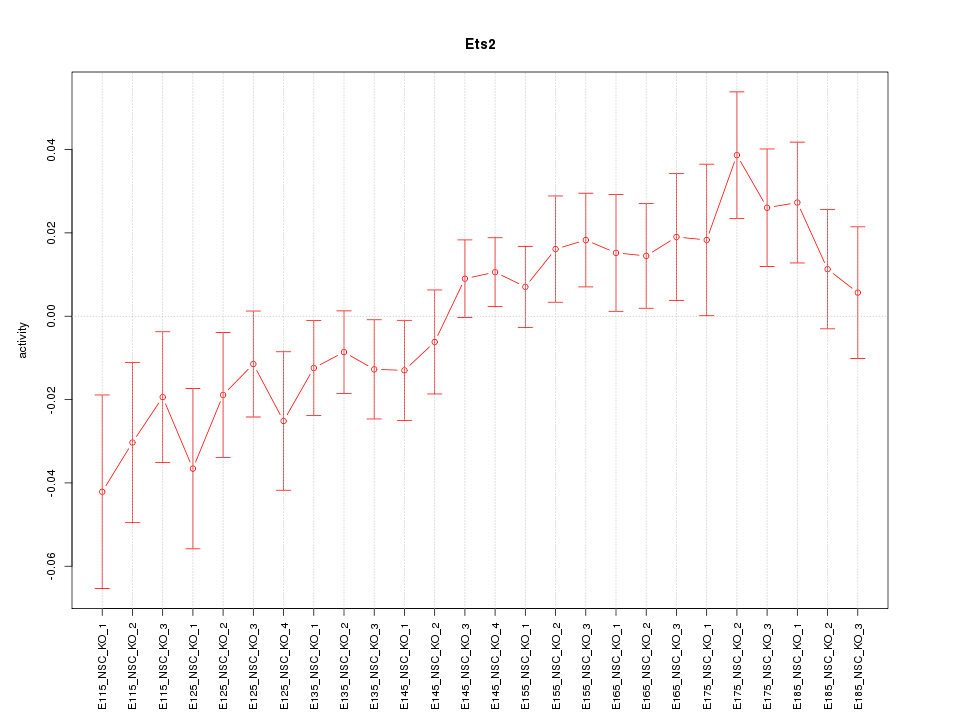
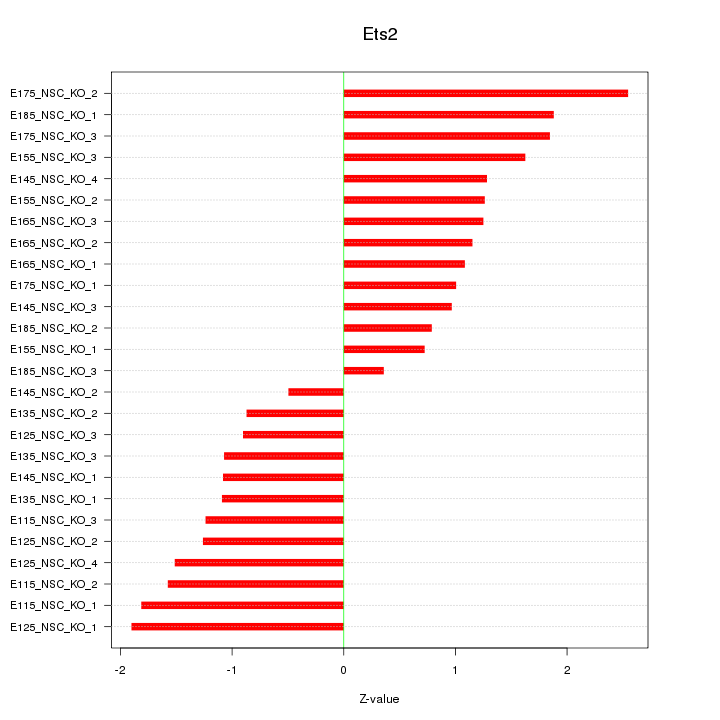
| 1.0 |
3.0 |
GO:0019085 |
early viral transcription(GO:0019085) |
| 0.8 |
8.0 |
GO:0090232 |
positive regulation of spindle checkpoint(GO:0090232) |
| 0.8 |
3.1 |
GO:0060715 |
syncytiotrophoblast cell differentiation involved in labyrinthine layer development(GO:0060715) |
| 0.7 |
10.5 |
GO:0021902 |
commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.6 |
3.7 |
GO:1902847 |
macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of tau-protein kinase activity(GO:1902949) |
| 0.6 |
1.2 |
GO:1901668 |
regulation of superoxide dismutase activity(GO:1901668) |
| 0.6 |
1.7 |
GO:0019043 |
positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) establishment of viral latency(GO:0019043) |
| 0.6 |
2.8 |
GO:0030259 |
lipid glycosylation(GO:0030259) |
| 0.5 |
4.3 |
GO:0002281 |
macrophage activation involved in immune response(GO:0002281) |
| 0.5 |
3.7 |
GO:0060267 |
positive regulation of respiratory burst(GO:0060267) |
| 0.5 |
1.8 |
GO:0018307 |
enzyme active site formation(GO:0018307) |
| 0.4 |
3.8 |
GO:0061088 |
sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.4 |
0.8 |
GO:0019086 |
late viral transcription(GO:0019086) |
| 0.4 |
1.6 |
GO:0003430 |
growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.4 |
21.1 |
GO:0047496 |
vesicle transport along microtubule(GO:0047496) |
| 0.4 |
1.6 |
GO:0010637 |
negative regulation of mitochondrial fusion(GO:0010637) |
| 0.4 |
3.0 |
GO:0070447 |
positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 0.4 |
1.9 |
GO:0071787 |
endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.4 |
2.2 |
GO:0043654 |
recognition of apoptotic cell(GO:0043654) |
| 0.4 |
1.1 |
GO:0007228 |
positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.4 |
1.4 |
GO:0042364 |
water-soluble vitamin biosynthetic process(GO:0042364) positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.4 |
1.4 |
GO:1904425 |
negative regulation of GTP binding(GO:1904425) |
| 0.3 |
3.1 |
GO:0036371 |
protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.3 |
1.7 |
GO:0015871 |
choline transport(GO:0015871) regulation of resting membrane potential(GO:0060075) |
| 0.3 |
3.0 |
GO:0018095 |
protein polyglutamylation(GO:0018095) |
| 0.3 |
2.7 |
GO:0036506 |
maintenance of unfolded protein(GO:0036506) protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.3 |
1.0 |
GO:0036092 |
phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.3 |
1.0 |
GO:0046122 |
purine deoxyribonucleoside metabolic process(GO:0046122) |
| 0.3 |
3.2 |
GO:0048149 |
behavioral response to ethanol(GO:0048149) |
| 0.3 |
0.6 |
GO:0035990 |
tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.3 |
1.5 |
GO:0002457 |
T cell antigen processing and presentation(GO:0002457) |
| 0.3 |
2.8 |
GO:1904153 |
negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.3 |
0.8 |
GO:0006624 |
vacuolar protein processing(GO:0006624) |
| 0.3 |
1.0 |
GO:1902774 |
late endosome to lysosome transport(GO:1902774) |
| 0.2 |
1.0 |
GO:0086042 |
cardiac muscle cell-cardiac muscle cell adhesion(GO:0086042) |
| 0.2 |
1.0 |
GO:1902255 |
positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902255) |
| 0.2 |
1.4 |
GO:0070166 |
enamel mineralization(GO:0070166) |
| 0.2 |
4.0 |
GO:0045792 |
negative regulation of cell size(GO:0045792) |
| 0.2 |
0.7 |
GO:0008612 |
peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.2 |
0.9 |
GO:0045053 |
protein retention in Golgi apparatus(GO:0045053) |
| 0.2 |
1.1 |
GO:0090164 |
asymmetric Golgi ribbon formation(GO:0090164) |
| 0.2 |
1.7 |
GO:1901727 |
positive regulation of histone deacetylase activity(GO:1901727) |
| 0.2 |
0.6 |
GO:1902277 |
negative regulation of pancreatic amylase secretion(GO:1902277) |
| 0.2 |
0.6 |
GO:1903774 |
positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.2 |
1.3 |
GO:0006123 |
mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.2 |
1.2 |
GO:0048102 |
autophagic cell death(GO:0048102) |
| 0.2 |
0.6 |
GO:1901552 |
positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.2 |
1.3 |
GO:0017196 |
N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.2 |
9.1 |
GO:0036465 |
synaptic vesicle recycling(GO:0036465) |
| 0.2 |
0.7 |
GO:1903215 |
regulation of mRNA modification(GO:0090365) negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.2 |
2.7 |
GO:0060363 |
cranial suture morphogenesis(GO:0060363) |
| 0.2 |
6.3 |
GO:0072583 |
clathrin-mediated endocytosis(GO:0072583) |
| 0.2 |
0.7 |
GO:0019355 |
nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.2 |
3.0 |
GO:0035235 |
ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.2 |
2.2 |
GO:1901898 |
negative regulation of relaxation of muscle(GO:1901078) regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.2 |
1.0 |
GO:0044034 |
negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.2 |
1.3 |
GO:0035635 |
entry of bacterium into host cell(GO:0035635) regulation of entry of bacterium into host cell(GO:2000535) |
| 0.2 |
1.9 |
GO:0034497 |
protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.2 |
0.8 |
GO:0070863 |
positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.1 |
0.9 |
GO:0035469 |
determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.1 |
0.4 |
GO:0070488 |
neutrophil aggregation(GO:0070488) |
| 0.1 |
1.7 |
GO:0000185 |
activation of MAPKKK activity(GO:0000185) |
| 0.1 |
1.4 |
GO:0071578 |
zinc II ion transmembrane import(GO:0071578) |
| 0.1 |
1.7 |
GO:2000096 |
positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.1 |
0.3 |
GO:0090327 |
negative regulation of locomotion involved in locomotory behavior(GO:0090327) |
| 0.1 |
2.6 |
GO:0007095 |
mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.1 |
2.1 |
GO:0060213 |
regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.1 |
0.5 |
GO:0010936 |
negative regulation of macrophage cytokine production(GO:0010936) |
| 0.1 |
4.1 |
GO:0015991 |
energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.1 |
0.6 |
GO:0015867 |
ATP transport(GO:0015867) |
| 0.1 |
0.4 |
GO:0043201 |
response to leucine(GO:0043201) cellular response to leucine(GO:0071233) |
| 0.1 |
0.4 |
GO:1902373 |
negative regulation of mRNA catabolic process(GO:1902373) regulation of selenocysteine incorporation(GO:1904569) regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.1 |
4.1 |
GO:0034314 |
Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.1 |
0.4 |
GO:1904431 |
positive regulation of t-circle formation(GO:1904431) |
| 0.1 |
0.4 |
GO:0010248 |
B cell negative selection(GO:0002352) establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.1 |
1.0 |
GO:0071044 |
histone mRNA catabolic process(GO:0071044) |
| 0.1 |
0.7 |
GO:0071712 |
ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.1 |
1.1 |
GO:0030049 |
muscle filament sliding(GO:0030049) |
| 0.1 |
1.2 |
GO:0006622 |
protein targeting to lysosome(GO:0006622) |
| 0.1 |
0.9 |
GO:0003073 |
regulation of systemic arterial blood pressure(GO:0003073) |
| 0.1 |
0.8 |
GO:0097039 |
protein linear polyubiquitination(GO:0097039) |
| 0.1 |
1.5 |
GO:0090161 |
Golgi ribbon formation(GO:0090161) |
| 0.1 |
3.5 |
GO:0018279 |
protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.1 |
2.4 |
GO:0061099 |
negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.1 |
0.8 |
GO:0030578 |
PML body organization(GO:0030578) |
| 0.1 |
0.8 |
GO:0045842 |
positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.1 |
0.1 |
GO:0010796 |
regulation of multivesicular body size(GO:0010796) |
| 0.1 |
1.1 |
GO:0006829 |
zinc II ion transport(GO:0006829) |
| 0.1 |
1.6 |
GO:0019835 |
cytolysis(GO:0019835) |
| 0.1 |
1.9 |
GO:0045725 |
positive regulation of glycogen biosynthetic process(GO:0045725) |
| 0.1 |
3.9 |
GO:0015914 |
phospholipid transport(GO:0015914) |
| 0.1 |
17.3 |
GO:0007266 |
Rho protein signal transduction(GO:0007266) |
| 0.1 |
0.5 |
GO:1990564 |
protein ufmylation(GO:0071569) protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.1 |
1.5 |
GO:0010669 |
epithelial structure maintenance(GO:0010669) |
| 0.1 |
0.3 |
GO:0050861 |
positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.1 |
0.2 |
GO:0010726 |
positive regulation of hydrogen peroxide metabolic process(GO:0010726) |
| 0.1 |
0.2 |
GO:0042117 |
monocyte activation(GO:0042117) |
| 0.1 |
0.5 |
GO:0045213 |
neurotransmitter receptor metabolic process(GO:0045213) |
| 0.1 |
1.2 |
GO:1900454 |
positive regulation of long term synaptic depression(GO:1900454) |
| 0.1 |
3.9 |
GO:0007585 |
respiratory gaseous exchange(GO:0007585) |
| 0.1 |
1.1 |
GO:1902287 |
semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.1 |
0.4 |
GO:0016255 |
attachment of GPI anchor to protein(GO:0016255) |
| 0.1 |
0.8 |
GO:0030157 |
pancreatic juice secretion(GO:0030157) |
| 0.1 |
1.0 |
GO:0071801 |
regulation of podosome assembly(GO:0071801) |
| 0.1 |
0.1 |
GO:0043323 |
positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.1 |
0.5 |
GO:0048268 |
clathrin coat assembly(GO:0048268) |
| 0.1 |
0.6 |
GO:0046415 |
urate metabolic process(GO:0046415) |
| 0.1 |
0.2 |
GO:0070070 |
proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.1 |
0.5 |
GO:0001771 |
immunological synapse formation(GO:0001771) |
| 0.1 |
0.8 |
GO:0071257 |
cellular response to electrical stimulus(GO:0071257) |
| 0.1 |
4.1 |
GO:0060976 |
coronary vasculature development(GO:0060976) |
| 0.1 |
1.2 |
GO:0045724 |
positive regulation of cilium assembly(GO:0045724) |
| 0.1 |
1.2 |
GO:0000731 |
DNA synthesis involved in DNA repair(GO:0000731) |
| 0.1 |
0.8 |
GO:0014067 |
negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.1 |
0.7 |
GO:0006895 |
Golgi to endosome transport(GO:0006895) |
| 0.1 |
0.3 |
GO:1902455 |
negative regulation of stem cell population maintenance(GO:1902455) |
| 0.1 |
0.9 |
GO:0009072 |
aromatic amino acid family metabolic process(GO:0009072) |
| 0.1 |
0.8 |
GO:2000505 |
regulation of energy homeostasis(GO:2000505) |
| 0.1 |
0.1 |
GO:0060263 |
regulation of respiratory burst(GO:0060263) |
| 0.1 |
0.4 |
GO:0006102 |
isocitrate metabolic process(GO:0006102) |
| 0.1 |
0.5 |
GO:0035871 |
protein K11-linked deubiquitination(GO:0035871) |
| 0.1 |
0.3 |
GO:1901655 |
cellular response to ketone(GO:1901655) |
| 0.1 |
0.2 |
GO:0048304 |
positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.1 |
0.3 |
GO:0050651 |
dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.1 |
0.3 |
GO:0006172 |
ADP biosynthetic process(GO:0006172) |
| 0.1 |
0.2 |
GO:0034241 |
positive regulation of macrophage fusion(GO:0034241) regulation of osteoclast proliferation(GO:0090289) |
| 0.1 |
2.7 |
GO:1902850 |
mitotic spindle assembly(GO:0090307) microtubule cytoskeleton organization involved in mitosis(GO:1902850) |
| 0.0 |
0.2 |
GO:0071361 |
cellular response to ethanol(GO:0071361) |
| 0.0 |
0.2 |
GO:0045896 |
regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 |
0.5 |
GO:0051352 |
negative regulation of ligase activity(GO:0051352) negative regulation of ubiquitin-protein transferase activity(GO:0051444) |
| 0.0 |
0.6 |
GO:0043649 |
dicarboxylic acid catabolic process(GO:0043649) |
| 0.0 |
0.5 |
GO:0021819 |
layer formation in cerebral cortex(GO:0021819) |
| 0.0 |
0.2 |
GO:0017183 |
peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 |
0.3 |
GO:0006977 |
DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.0 |
1.2 |
GO:0050873 |
brown fat cell differentiation(GO:0050873) |
| 0.0 |
1.3 |
GO:0070306 |
lens morphogenesis in camera-type eye(GO:0002089) lens fiber cell differentiation(GO:0070306) |
| 0.0 |
1.1 |
GO:0046854 |
phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 |
0.5 |
GO:0070234 |
positive regulation of T cell apoptotic process(GO:0070234) |
| 0.0 |
0.1 |
GO:1903378 |
positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.0 |
2.0 |
GO:0060395 |
SMAD protein signal transduction(GO:0060395) |
| 0.0 |
1.4 |
GO:1901998 |
toxin transport(GO:1901998) |
| 0.0 |
0.7 |
GO:0006656 |
phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.0 |
0.1 |
GO:0006436 |
tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.0 |
0.3 |
GO:0031848 |
protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 |
0.1 |
GO:0090370 |
negative regulation of cholesterol efflux(GO:0090370) |
| 0.0 |
0.3 |
GO:0019363 |
pyridine nucleotide biosynthetic process(GO:0019363) |
| 0.0 |
0.5 |
GO:0006491 |
N-glycan processing(GO:0006491) |
| 0.0 |
0.5 |
GO:0097576 |
autophagosome maturation(GO:0097352) vacuole fusion(GO:0097576) |
| 0.0 |
0.7 |
GO:0010800 |
positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 |
0.5 |
GO:0015012 |
heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.0 |
0.7 |
GO:0045879 |
negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.0 |
0.7 |
GO:0008333 |
endosome to lysosome transport(GO:0008333) |
| 0.0 |
1.1 |
GO:0071526 |
semaphorin-plexin signaling pathway(GO:0071526) |
| 0.0 |
0.3 |
GO:0006903 |
vesicle targeting(GO:0006903) |
| 0.0 |
0.7 |
GO:0035066 |
positive regulation of histone acetylation(GO:0035066) |
| 0.0 |
1.1 |
GO:2000649 |
regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 |
0.1 |
GO:0045041 |
protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.0 |
1.9 |
GO:0032091 |
negative regulation of protein binding(GO:0032091) |
| 0.0 |
0.4 |
GO:0032400 |
melanosome localization(GO:0032400) |
| 0.0 |
0.1 |
GO:0045650 |
negative regulation of macrophage differentiation(GO:0045650) |
| 0.0 |
2.8 |
GO:0010977 |
negative regulation of neuron projection development(GO:0010977) |
| 0.0 |
1.2 |
GO:0042147 |
retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 |
1.3 |
GO:2001238 |
positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.0 |
1.1 |
GO:0035019 |
somatic stem cell population maintenance(GO:0035019) |
| 0.0 |
0.3 |
GO:0010719 |
negative regulation of epithelial to mesenchymal transition(GO:0010719) |
| 0.0 |
5.8 |
GO:0043087 |
regulation of GTPase activity(GO:0043087) |
| 0.0 |
0.3 |
GO:0000301 |
retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 |
0.1 |
GO:0031442 |
positive regulation of mRNA 3'-end processing(GO:0031442) |
| 0.0 |
0.2 |
GO:0071218 |
cellular response to misfolded protein(GO:0071218) |
| 0.0 |
2.1 |
GO:0030010 |
establishment of cell polarity(GO:0030010) |
| 0.0 |
0.4 |
GO:0016558 |
protein import into peroxisome matrix(GO:0016558) |
| 0.0 |
0.9 |
GO:0006376 |
mRNA splice site selection(GO:0006376) |
| 0.0 |
0.7 |
GO:0031146 |
SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 |
1.1 |
GO:0045665 |
negative regulation of neuron differentiation(GO:0045665) |
| 0.0 |
0.5 |
GO:0000184 |
nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 |
0.1 |
GO:0034472 |
snRNA 3'-end processing(GO:0034472) |
| 0.0 |
0.1 |
GO:0006620 |
posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 |
0.1 |
GO:0048742 |
regulation of skeletal muscle fiber development(GO:0048742) |
| 0.0 |
1.8 |
GO:0098656 |
anion transmembrane transport(GO:0098656) |
| 0.0 |
0.6 |
GO:0045921 |
positive regulation of exocytosis(GO:0045921) |
| 0.0 |
0.1 |
GO:1900095 |
regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.0 |
0.1 |
GO:0044804 |
nucleophagy(GO:0044804) |
| 0.0 |
0.5 |
GO:0045761 |
regulation of adenylate cyclase activity(GO:0045761) |
| 0.0 |
1.3 |
GO:0016579 |
protein deubiquitination(GO:0016579) |
| 0.0 |
0.2 |
GO:0032981 |
NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 |
0.0 |
GO:0090394 |
negative regulation of excitatory postsynaptic potential(GO:0090394) |